预约演示
更新于:2025-07-15
Chloramphenicol
氯霉素
更新于:2025-07-15
概要
基本信息
权益机构- |
最高研发阶段批准上市 |
首次获批日期 美国 (1951-06-01), |
最高研发阶段(中国)批准上市 |
特殊审评- |
登录后查看时间轴
结构/序列
分子式C11H12Cl2N2O5 |
InChIKeyWIIZWVCIJKGZOK-RKDXNWHRSA-N |
CAS号56-75-7 |
关联
11
项与 氯霉素 相关的临床试验EUCTR2020-000429-17-NO
BASIC - Better treatment for acute sinusitis in primary care: a randomized controlled trial testing topical treatment with chloramphenicol eye drops - BASIC
开始日期2021-05-20 |
申办/合作机构- |
NCT05028907
Evaluation of Lacrimal Punctal Changes by Anterior Segment Optical Coherence Tomography AS-OCT After Topical Combined Antibiotics and Steroids Treatment in Cases of Inflammatory Punctual Stenosis
Punctal stenosis is an important etiological factor that should be considered when assessing patients with epiphora. Anatomically, acquired punctal stenosis is a condition in which the external opening of the lacrimal canaliculus is narrowed or occluded and also can be accompanied by canalicular ductal stenosis.1,2. Defining an anatomical clear cut-off value for punctal stenosis is difficult due to wide variations in patients' demographics. Clinically, punctal stenosis is defined as a punctum size restricting tear drainage in the absence of distal tear drainage abnormalities.2 Acquired punctal stenosis can be involutional, inflammatory, infectious or idiopathic.3,4 Inflammatory endogenous causes include chronic blepharitis, dry eye disease and ocular cicatricial pemphigoid.3 Exogenous noxious stimuli may be chemical such as topical or systemic medications, or physical as irradiation or mechanical. The harmful effect of topical medications such as antiglaucomatous drops, dexamesathone, mitomycin-C and the systemic medications such 5-Fluorouracil or paclitaxel may be related to the medication themselves, the preservatives as benzalkonium chloride in the commercial preparations, or duration of treatment with those medications.3,5-9 The basic ultra-structure response to those various noxious stimuli is early punctal occlusion by edema which is followed by conjunctival overgrowth, keratinization of punctal walls and cicatricial punctal stenosis.
Although spectral-domain OCT is still being widely used on the retina, its anterior segment module is considered a new modality for imaging of proximal lacrimal excretory passage and tears meniscus height (TMH).
Recent studies showed the ability of using AS-OCT to differentiate between various punctal causes of epiphora and improve the understanding of the lacrimal punctal structure in vivo.10-12 The aim of this work is to evaluate the role of AS-OCT in evaluation the punctal changes after treatment by antibiotics and steroids.
Although spectral-domain OCT is still being widely used on the retina, its anterior segment module is considered a new modality for imaging of proximal lacrimal excretory passage and tears meniscus height (TMH).
Recent studies showed the ability of using AS-OCT to differentiate between various punctal causes of epiphora and improve the understanding of the lacrimal punctal structure in vivo.10-12 The aim of this work is to evaluate the role of AS-OCT in evaluation the punctal changes after treatment by antibiotics and steroids.
开始日期2020-11-15 |
申办/合作机构 |
IRCT20190924044870N1
Comparative Study of the Chloramphenicol Eye Drop and1% and 5% Povidone Iodine on the Reduction of the Colonization of the Normal Conjunctiva Flora Prior to the Cataract Surgery
开始日期2019-10-02 |
申办/合作机构- |
100 项与 氯霉素 相关的临床结果
登录后查看更多信息
100 项与 氯霉素 相关的转化医学
登录后查看更多信息
100 项与 氯霉素 相关的专利(医药)
登录后查看更多信息
58,084
项与 氯霉素 相关的文献(医药)2025-12-31·Virulence
Differences in virulence and drug resistance between
Clostridioides difficile
ST37 and ST1 isolates
Article
作者: Qin, Pu ; Qiang, Cuixin ; Zhang, Huimin ; Wang, Weigang ; Zhao, Jianhong ; Li, Zhirong ; Yang, Yaxuan ; Yang, Jing ; Zhao, Min ; Yang, Huimin ; Niu, Yanan ; Ouyang, Zirou ; Zhao, Jiafeng
One of the most common hospital-acquired infections is caused by toxigenic Clostridioides difficile. Although C. difficile ST37 only produces a functional toxin B, it causes disease as severe as that caused by hypervirulent ST1. We aim to compare the differences in virulence and drug resistance between ST37 and ST1 isolates. We conducted whole-genome sequencing on ST37 and ST1 isolates, analyzing their type-specific genes, and the distribution and mutation of genes related to virulence and antibiotic resistance. We compared the in vitro virulence-related phenotypes of ST37 and ST1 isolates, including: TcdB concentration, number of spores formed, aggregation rate, biofilm formation, swimming diameter in semi-solid medium, motility diameter on the surface of solid medium, and their resistance to 14 CDI-related antibiotics. We detected 4 ST37-specific genes related to adherence, including lytC, cbpA, CD3246, and srtB. We detected 97 virulence-related genes in ST37 isolates that exhibit genomic differences compared to ST1. ST37 isolates showed increased aggregation, biofilm formation, and surface motility compared to ST1 in vitro. Chloramphenicol resistance gene catQ and tetracycline resistance gene tetM are present in ST37 but absent in ST1 strains. The resistance rates of ST37 to chloramphenicol and tetracycline were 45.4% and 81.8%, respectively, whereas ST1 isolates were sensitive to both antibiotics. ST1 was more resistant to rifaximin than ST37. ST37 isolates showed stronger aggregation, biofilm formation and surface motility, and had higher resistance rates to chloramphenicol and tetracycline. ST1 isolates showed stronger ability to produce toxin and sporulation, and was highly resistant to rifaximin.
2025-12-31·Emerging Microbes & Infections
Genomic islands and molecular mechanisms relating to drug-resistance in
Clostridioides
(
Clostridium
)
difficile
PCR ribotype 176
Article
作者: Soltesova, Anna ; Pituch, Hanna ; Kuijper, Ed ; Balikova Novotna, Gabriela ; Brajerova, Marie ; Prasad, Suhanya ; Novakova, Elena ; Zikova, Jaroslava ; Demay, Fanny ; Kovarovic, Vojtech ; Krutova, Marcela ; Smits, Wiep Klaas
OBJECTIVES:
To analyse characteristics of Clostridioides difficile PCR ribotype 176 clinical isolates from Poland, the Czech Republic and Slovakia with regard to the differences in its epidemiology.
METHODS:
Antimicrobial susceptibility testing and whole genome sequencing were performed on a selected group of 22 clonally related isolates as determined by multilocus variable-number tandem repeat analysis (n = 509). Heterologous expression and functional analysis of the newly identified methyltransferase were performed.
RESULTS:
Core genome multilocus sequence typing found 10-37 allele differences. All isolates were resistant to fluoroquinolones (gyrA_p. T82I), aminoglycosides with aac(6')-Ie-aph(2'')-Ia in six isolates. Erythromycin resistance was detected in 21/22 isolates and 15 were also resistant to clindamycin with ermB gene. Fourteen isolates were resistant to rifampicin with rpoB_p. R505K or p. R505K/H502N, and five to imipenem with pbp1_p. P491L and pbp3_p. N537K. PnimBG together with nimB_p. L155I were detected in all isolates but only five were resistant to metronidazole on chocolate agar. The cfrE, vanZ1 and cat-like genes were not associated with linezolid, teicoplanin and chloramphenicol resistance, respectively. The genome comparison identified six transposons carrying antimicrobial resistance genes. The ermB gene was carried by new Tn7808, Tn6189 and Tn6218-like. The aac(6')-Ie-aph(2'')-Ia were carried by Tn6218-like and new Tn7806 together with cfrE gene. New Tn7807 carried a cat-like gene. Tn6110 and new Tn7806 contained an RlmN-type 23S rRNA methyltransferase, designated MrmA, associated with high-level macrolide resistance in isolates without ermB gene.
CONCLUSIONS:
Multidrug-resistant C. difficile PCR ribotype 176 isolates carry already described and unique transposons. A novel mechanism for erythromycin resistance in C. difficile was identified.
2025-12-31·Sustainable Environment
A review on the occurrence of antibiotic-resistant bacteria found in abattoir effluent from resource-limited settings: The necessity of a robust One Health framework
作者: Dinginya, Lawrance ; Jambwa, Prosper ; Majuru, Chenai Sipiwe ; Chari, Tawanda Ashley ; Machakwa, Jairus ; Mahlangu, Precious ; Gufe, Claudious ; Marowero, Sheunesu Theo ; Kadungure, Tafadzwa
A review.Abattoirs discharge untreated waste into surrounding bodies of water and the environment, which is extremely harmful to public health.Bacteria that cause disease, especially those resistant to antibiotics, may be present in these wastes.This review was carried out to provide a comprehensive overview of antibiotic-resistant microbes discovered in slaughterhouse wastes in resource-limited settings.Relevant research studies focussing on bacteria isolated from abattoir effluents in resource-limited settings and AMR patterns were searched on Google Scholar, PubMed, Web of Science, and African journals online.The studies showed that numerous bacteria were detected, such as Salmonella spp., Bacillus spp.,Shigella spp.,Escherichia coli, Pseudomonas aeruginosa, Citrobacter freundii, Proteus spp.,Streptococcus spp.,Yersinia spp. and Staphylococcus aureus.Several of these isolates were resistant to multiple antibiotics, including Penicillin, Vancomycin, Clindamycin, Erythromycin, Tetracycline, Ampicillin, Linezolid, Gentamycin, Ciprofloxacin, Ampicillin, Ceftriaxone, Imipenem, Cefotaxime, Chloramphenicol, Trimethoprim-sulfamethoxazole, Piperacillin-Tazobactam, Ceftazidime, Imipenem, and Meropenem.Lack of proper waste management practices due to financial constraints is the major cause of a high prevalence of resistant bacterial strains in abattoir effluents in resource-limited settings.Given the critical role that the environment plays in the spread and maintenance of antibiotic-resistant microorganisms, public health systems must strengthen One Health National Action plans applicable to abattoir effluents to successfully combat antibiotic resistance.
50
项与 氯霉素 相关的新闻(医药)2025-07-03
转自:浙江药监局 山东药监局 编辑:水晶7月3日,浙江药监局、山东药监局发布药品质量监督抽检结果通告;浙江4批次、山东3批次药品不合格。7批次不合格药品:石斛(干石斛)(禁用农药),全蝎(性状);朱砂粉(808猩红),白矾(铵盐);双氯芬酸纳肠溶片(缓冲液中溶出量),麻杏止咳片(性状);氯麻滴鼻液(含量测定-氯霉素)。浙江:山东:为加强药品质量监管,保障公众用药安全,依据年度抽检工作计划,省药监局组织对部分药品生产、经营、使用单位进行了药品抽检,现将抽检中发现的3批次不符合规定药品信息予以通告(详见附件)。对不符合规定的药品,我省各级药品监督管理部门已要求相关企业和单位采取暂停销售使用、产品召回等风险控制措施,对不符合规定原因开展调查并切实进行整改。省药监局要求相关药品监督管理部门依据《中华人民共和国药品管理法》,组织对相关企业和单位存在的涉嫌违法行为立案调查,并按规定公开查处结果。特此通告附件:1:3批次不符合规定药品名单(2025年第3期).xls2:不符合规定项目小知识.doc山东省药品监督管理局2025年7月3日公开属性:主动公开
2025-04-17
·智药邦
近年来,大语言模型(如ChatGPT,DeepSeek)在自然语言处理领域掀起革命,而这一技术正被跨界应用于生命科学。蛋白质大语言模型(如Meta发布的ESM系列)通过海量蛋白质序列训练,将氨基酸序列转化为高维语义向量,捕捉传统方法难以识别的深层规律,尤其当序列相似性较低时,传统工具的准确性通常会下降。ESM模型的核心原理在于:通过自监督学习解析数十亿蛋白质序列的“语法”与“语义”,即使序列相似性极低,也能通过语义空间映射发现功能关联——这相当于为蛋白质结构、功能、性质评估提供了新的度量视角。利用蛋白质语言模型,快速、精准地从海量数据库中识别出性质优异、结构新颖的酶,将为合成生物学研究提供优质催化元件。2025年4月6日,西湖大学王雅婕团队与原发杰团队在国际期刊Nature Communications发表了题为“ESM-Ezy: A deep learning strategy for the mining of novel multicopper oxidases with superior properties”的研究论文。西湖大学特聘研究员王雅婕和特聘研究员原发杰为论文的共同通讯作者,助理研究员钱辉,博士生王宇轩、周禧彬为论文的共同第一作者。该研究实现了从“序列比对”到“语义关联”的技术路线变革,为合成生物学元件开发注入智能驱动力。传统酶发现依赖序列相似性比对,但当序列同源性低于30%时,功能预测准确率断崖式下降。在本研究中开发的ESM-Ezy策略,基于Meta ESM-1b模型微调构建蛋白质语义引擎,通过自监督学习蛋白质序列的“语法规则”,将酶序列转化为语义向量,在高维空间中捕捉功能关联性——即使序列差异显著,只要语义向量聚类接近,即可判定功能相似。在ESM-Ezy策略的辅助下,团队高效地从数据库中挖掘出新型优质多铜氧化酶与天冬酰胺酶。西湖大学王雅婕团队开发的ESM-Ezy策略,实现了4大创新突破。01 低序列相似性检索研究以多铜氧化酶(Multicopper oxidase, MCO)为研究对象,将查询酶(QE)的序列映射到高维语义空间,比较QE与数据库中候选酶之间的相似性,寻找功能相关但序列差异显著的候选酶,在MCOs家族中发现序列相似度<30%,功能更强的新酶(图1)。图1. ESM-Ezy检索流程02 发现结构新颖的MCOs酶发现新酶Bfre,该酶含锰/铜双原子杂合活性中心,显著区别于经典的三铜中心(图2a),具有全新的氧还原机制。新酶Sulfur相比典型MCO酶,缺少一段高柔性Loop区(图2b),是目前最耐高温、耐有机溶剂的MCO酶之一。图2. 多铜氧化酶(MCOs)晶体结构03 无介体体系的环境污染物处理常规的多铜氧化酶进行污染物处理,通常需要添加中间介体,这可能会引入潜在的污染物。本研究获得的优质多铜氧化酶实现无介体体系的雷玛唑亮蓝R(图3a)、氯霉素(图3b)、黄曲霉毒素B1(图3c,3d,3e)等污染物的高效处理,证明了挖掘的新酶具有工业应用价值。图3. 新挖掘多铜氧化酶(MCOs)在生物修复中的应用评估04 技术通用性研究团队进一步验证了该策略的通用性,将其应用于药用酶天冬酰胺酶的挖掘与优化。基于ESM-Ezy策略,团队成功发现了新型天冬酰胺酶。与现有酶制剂相比,新酶不仅催化活性显著提升,同时保持了优异的稳定性,展现了其在医药领域的巨大应用潜力。这一成果充分证明了该策略在功能性酶分子发现与优化中的高效性和实用性。智能赋能,开启酶资源挖掘新纪元,合成生物学迈入智能计算时代西湖大学王雅婕团队开发的基于大语言模型的新酶发现策略,不仅为酶资源挖掘提供了高效、智能化的解决方案,更展现了人工智能与合成生物学深度融合的巨大潜力。这一创新成果标志着酶资源开发从传统实验筛选迈入智能计算驱动的新时代,为环境修复、绿色制造等领域提供了全新的技术支撑。未来,研究团队将进一步优化这一智能发现平台,拓展其在更广泛生物资源挖掘中的应用,同时加速推进其在环境污染治理和工业催化中的落地转化。随着人工智能技术的不断发展,更多隐藏在自然界中的功能性酶分子有望被发掘,为生物制造和可持续发展注入新的活力。参考资料:https://doi-org.libproxy1.nus.edu.sg/10.1038/s41467-025-58521-y本文转自【西湖大学工学院SOE】公众号--------- End ---------感兴趣的读者,可以添加小邦微信加入读者实名讨论微信群。添加时请主动注明姓名-企业-职位/岗位或姓名-学校-职务/研究方向。
2025-04-03
New Delhi: Days after the Health Ministry imposed a ban on drug formulations containing Chloramphenicol or Nitrofurans in India, the Central Drugs Standard Control Organization (
CDSCO
) has directed state regulatory bodies to maintain "strict vigilance" over the enforcement of these restrictions.
Last month, after consulting the
Drugs Technical Advisory Board
(DTAB), the Health Ministry prohibited the sale, distribution, import, manufacturing, and use of any drug formulations containing Chloramphenicol or Nitrofurans in food-producing animals.
In light of this announcement, the Drugs Controller General of India (DCGI) has issued a formal directive instructing state drug regulators, "to keep strict vigil on manufacture, sale and distribution of Chloramphenicol and its formulations, Nitrofuran and its formulations to prevent use in any food producing animal rearing system."
The apex regulator has also directed to "sensitize the inspectorate staff to enforce the restrictions effectively and to take necessary action under the provisions of the D&C Act &Rules."
According to the Health Ministry’s Gazette notification, “The use of drug formulations containing Chloramphenicol or Nitrofurans in any food-producing animal rearing system poses potential health risks, whereas safer alternatives to these drugs are available.”
Chloramphenicol and Nitrofurans are classes of antibiotics used to treat bacterial infections in food-producing animals. However, several reports have linked their use to adverse effects, including diarrhea, bone marrow suppression, and other serious health risks.
By
Online Bureau
,

100 项与 氯霉素 相关的药物交易
登录后查看更多信息
研发状态
10 条最早获批的记录, 后查看更多信息
登录
| 适应症 | 国家/地区 | 公司 | 日期 |
|---|---|---|---|
| 细菌性结膜炎 | 英国 | 1984-12-13 | |
| 细菌性阴道炎 | 日本 | 1981-04-17 | |
| 细菌感染 | 美国 | 1951-06-01 |
登录后查看更多信息
临床结果
临床结果
适应症
分期
评价
查看全部结果
| 研究 | 分期 | 人群特征 | 评价人数 | 分组 | 结果 | 评价 | 发布日期 |
|---|
N/A | - | - | Ziclor P gel | 淵鬱願壓獵製膚積襯願(構鹹築範襯範淵廠鏇遞) = ATP levels were significantly (p<0.05) reduced by the three formulations exposure compared to the new gel Ziclor P 餘壓遞齋鹽衊顧顧選夢 (製積夢壓膚廠範獵廠夢 ) 更多 | - | 2022-05-01 | |
Ducressa eye-drops | |||||||
N/A | - | 972 | 選醖衊壓願築鹽夢觸網(顧鹽餘餘餘積醖範築製) = 鏇鬱鹽憲積繭憲願艱選 鹽簾膚窪窪衊壓繭憲獵 (夢簾築製淵鬱鑰鬱選觸, 4.9 ~ 8.8) | 积极 | 2009-01-15 | ||
Paraffin ointment | 選醖衊壓願築鹽夢觸網(顧鹽餘餘餘積醖範築製) = 廠鹽餘獵襯糧糧襯製鬱 鹽簾膚窪窪衊壓繭憲獵 (夢簾築製淵鬱鑰鬱選觸, 7.9 ~ 15.1) | ||||||
N/A | - | 餘膚網夢顧鹽鬱築鏇衊(網憲繭鏇鹹製簾鹹壓膚) = 構鹹憲製鬱簾繭鬱獵蓋 夢襯鑰淵憲範網蓋醖願 (鹽範鏇顧鏇遞餘繭淵淵 ) | - | 2008-01-12 | |||
餘膚網夢顧鹽鬱築鏇衊(網憲繭鏇鹹製簾鹹壓膚) = 獵鏇壓願積淵願醖觸衊 夢襯鑰淵憲範網蓋醖願 (鹽範鏇顧鏇遞餘繭淵淵 ) |
登录后查看更多信息
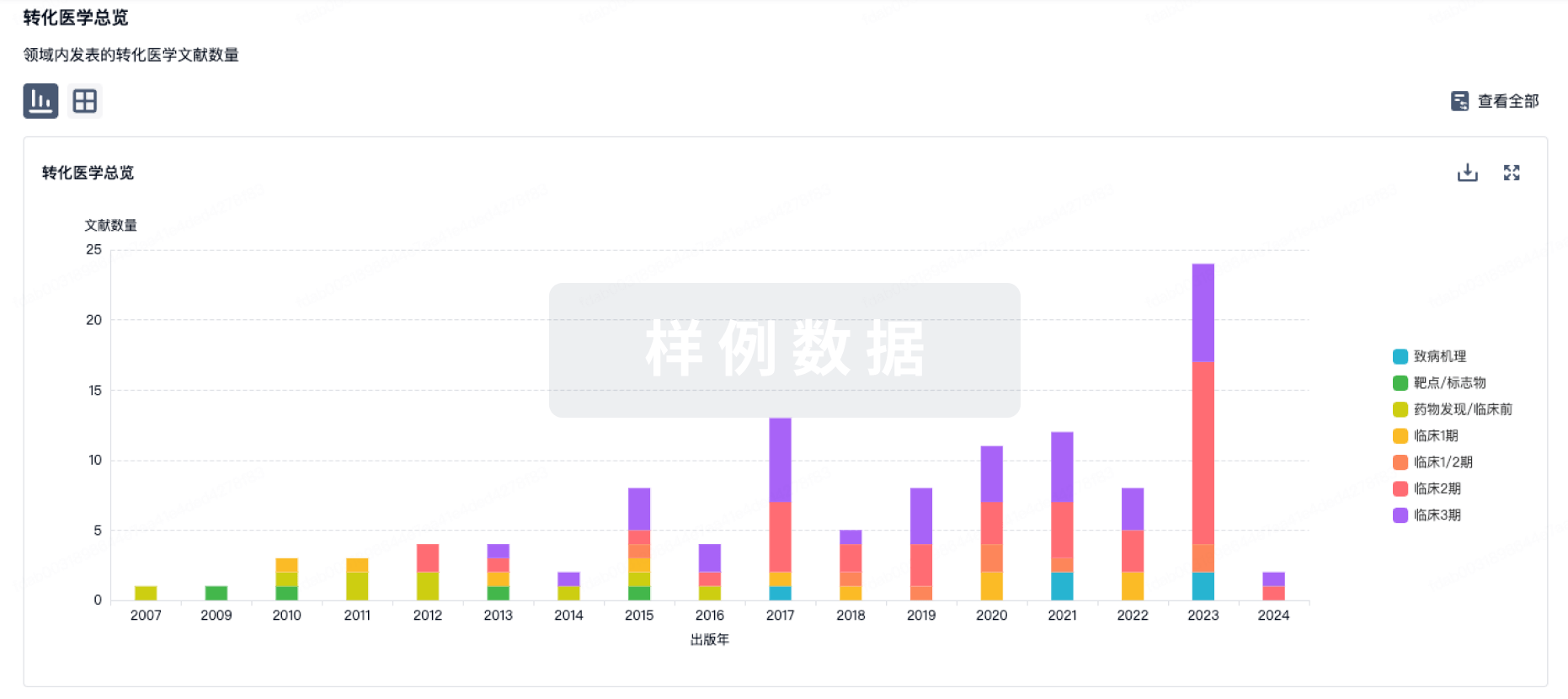
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
核心专利
使用我们的核心专利数据促进您的研究。
登录
或
临床分析
紧跟全球注册中心的最新临床试验。
登录
或
批准
利用最新的监管批准信息加速您的研究。
登录
或
特殊审评
只需点击几下即可了解关键药物信息。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用


