预约演示
更新于:2025-05-07
PARP1 x PARP2 x PARP
更新于:2025-05-07
关联
31
项与 PARP1 x PARP2 x PARP 相关的药物作用机制 PARP1抑制剂 [+1] |
在研机构 |
原研机构 |
最高研发阶段批准上市 |
首次获批国家/地区 中国 |
首次获批日期2025-01-14 |
作用机制 PARP1抑制剂 [+1] |
在研机构 |
原研机构 |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 中国 |
首次获批日期2021-04-30 |
1,159
项与 PARP1 x PARP2 x PARP 相关的临床试验NCT04982848
A Prospective, Single-arm, Open-label, Non-interventional, Multi-centre, Post Marketing Surveillance (PMS) Study of Talzenna(Registered)
Talzenna will be approved for the treatment of gBRCA advanced breast cancer in Korea. In accordance with the Standards for Re-examination of New Drug, it is required to conduct a PMS. Post marketing surveillance is required to determine any problems or questions associated with Talzenna after marketing in Korea, with regard to the following clauses under conditions of general clinical practice. Therefore, through this study, effectiveness and safety of Talzenna will be observed.
开始日期2025-12-31 |
申办/合作机构 |
NCT06856499
Phase I Evaluation of Combination CLK/DYRK (Cirtuvivint) Inhibition with PARP Inhibition (Olaparib) in BRCA/HRD Platinum Resistant Ovarian Cancer
The purpose of this study is to learn about the safety and tolerability of Cirtuvivint in combination with Olaparib in platinum resistant ovarian cancer. The study also aims to determine the recommended dose of the combination therapy.
If a participant is a good fit for the study, and they enroll in the study, they will:
* Visit the clinic often at the beginning of the study for physical exams, blood draws, vital signs, and other study and routine care procedures. After the first two months participants will visit the clinic every 28 days.
* Take the study medications, Cirtuvivint and Olaparib. Participants will take Olaparib every day. Participants will either take Cirtuvivint 5 days per week or 2 days per week.
If a participant is a good fit for the study, and they enroll in the study, they will:
* Visit the clinic often at the beginning of the study for physical exams, blood draws, vital signs, and other study and routine care procedures. After the first two months participants will visit the clinic every 28 days.
* Take the study medications, Cirtuvivint and Olaparib. Participants will take Olaparib every day. Participants will either take Cirtuvivint 5 days per week or 2 days per week.
开始日期2025-12-01 |
申办/合作机构 |
100 项与 PARP1 x PARP2 x PARP 相关的临床结果
登录后查看更多信息
100 项与 PARP1 x PARP2 x PARP 相关的转化医学
登录后查看更多信息
0 项与 PARP1 x PARP2 x PARP 相关的专利(医药)
登录后查看更多信息
447
项与 PARP1 x PARP2 x PARP 相关的文献(医药)2025-06-01·Bioorganic Chemistry
Engaging an engineered PARP-2 catalytic domain mutant to solve the complex structures harboring approved drugs for structure analyses
Article
作者: Xu, Bailing ; Wang, Xiaoyu ; Zhou, Jie
2025-05-01·DNA Repair
PARP inhibitors in ovarian cancer: Mechanisms of resistance and implications to therapy
Article
作者: Madhusudan, Srinivasan ; Kulkarni, Sanat ; Seneviratne, Nethmin ; Tosun, Çağla
2025-04-01·Biochemical Pharmacology
Parps in immune response: Potential targets for cancer immunotherapy
Review
作者: Chen, Yali ; Wang, Shuping ; Zeng, Tingyu ; Huang, Jingling ; Xu, Yungen ; Zhang, Bangzhi
125
项与 PARP1 x PARP2 x PARP 相关的新闻(医药)2025-04-20
据 Insight 数据库统计,本周(4 月 13 日—4 月 19 日)全球共有 59 款创新药(含改良新)研发进度推进到了新阶段,其中 2 款获批上市,1 款申报上市,8 款启动临床,16 款获批临床,18 款申报临床。下文,Insight 将分别摘取本周国内外部分重点项目进展做介绍。境外创新药进展境外部分,本周共有 13 款药物研发阶段推进,包括 1 款获批上市,1 款首次启动 I 期临床,9 款首次获批临床。 获批上市据 Insight 数据库显示,本周共有 7 条新药/新适应症在三大海外主要国家/地区(美国、EMA、日本)获批,详情如下:1、赛诺菲/再生元:「度普利尤单抗」获 FDA 批准新适应症当地时间 4 月 18 日,再生元和赛诺菲宣布,FDA 已批准度普利尤单抗(Dupilumab)的一项新适应症上市,用于治疗使用 H1 抗组胺药控制不佳的 12 岁及以上儿童和成人慢性自发性荨麻疹(CSU)。新闻稿指出,度普利尤单抗是美国十多年来首个针对 CSU 的靶向治疗药物。截图来源:赛诺菲官网度普利尤单抗由赛诺菲和再生元联合开发,是美国 FDA 批准的首个抗 IL-4Rα抗体。自 2017 年获批上市至今,度普利尤单抗已在 60 多个国家/地区范围内获批了多种炎症性疾病,包括特应性皮炎、哮喘、结节性痒疹、慢性阻塞性肺疾病、嗜酸性细胞性食管炎等。2024 年,度普利尤单抗全球销售额达到近 141 亿美元,同比增长 21.57%,成为了自免领域的新一代药王。本次批准基于 Ⅲ 期临床研究 LIBERTY-CUPID 的研究 A 和研究 C 的数据,两项研究均达到了其主要终点和关键次要终点。研究 B 提供了额外的安全性数据。LIBERTY-CUPID 研究的三部分研究具体数据如下。● 研究 A 纳入了 136 名受试者,评估了度普利尤单抗对比安慰剂治疗既往接受过抗组胺药(标准治疗)治疗但未接受过奥马珠单抗治疗的 6 岁及以上儿童和成人 CSU 患者的疗效和安全性。结果显示:对比安慰剂组,度普利尤单抗组的瘙痒严重评分显著降低(-10.2 vs. -6.0 分,p=0.0005);度普利尤单抗组的荨麻疹活动度评分也显著降低(-20.5 vs. -12.0 分,p=0.0003)。● 研究 B 纳入 108 名受试者,评估度普利尤单抗对比安慰剂治疗既往接受过抗组胺药治疗且对奥马珠单抗不耐受或不完全反应的 12-80 岁青少年和成人 CSU 患者的疗效和安全性。结果显示:相比安慰剂组,度普利尤单抗组的瘙痒严重评分降低(-7.7 vs. -4.8 分,p=0.0449),度普利尤单抗组的荨麻疹活动度评分降低(-14.2 vs. -8.5 分,p=0.0039),但统计学意义不够显著。● 研究 C 纳入 148 名受试者,评估了度普利尤单抗对比安慰剂治疗既往接受过抗组胺药治疗但未接受过奥马珠单抗治疗的 6 岁及以上儿童和成人 CSU 患者的疗效和安全性。结果显示:在 24 周时,与安慰剂相比,度普利尤单抗组的瘙痒严重评分显著降低(-8.64 vs. -6.10 分,p=0.02);荨麻疹活动度也显著降低(-15.86 vs. -11.21 分,p=0.02);疾病控制(荨麻疹活动评分≤6)患者比例更高(41% vs. 23%,p=0.05);完全缓解(荨麻疹活动评分=0)的患者比例也更高(30% vs. 18%,p=0.02)。安全性方面,所有 LIBERTY-CUPID 3 期研究的安全性结果与度普利尤单抗已知的安全性结果基本一致。与安慰剂相比,度普利尤单抗更常见的不良事件(≥5%)是注射部位反应和 COVID-19 感染。2、罗氏:CD20/CD3 双抗在欧盟获批新适应症当地时间 4 月 14 日,罗氏宣布格罗菲妥单抗(Glofitamab)联合吉西他滨和奥沙利铂(GemOx)组合疗法获 EMA 批准,用于治疗不适合接受自体干细胞移植(ASCT)的复发或难治性(R/R)弥漫大 B 细胞淋巴瘤(DLBCL)成年患者。此次批准后,格罗菲妥单抗的这一组合疗法成为欧洲首个可用于癌症复发或对初始治疗无反应的 DLBCL 患者的双特异性抗体治疗方案。截图来源:企业官网格菲妥单抗是一种 CD20/CD3 T 细胞结合双特异性抗体,据 Insight 数据库显示,该药于 23 年 6 月获 FDA 批准上市,用于 DLBCL 三线治疗,同年 6 月和 11 月分别获 EMA 和 NMPA 批准。此次批准基于关键性 III 期 STARGLO 研究的结果。这是一项在 R/R DLBCL 患者中评价格菲妥单抗联合 GemOx 对比利妥昔单抗联合 GemOx 的有效性和安全性的 III 期、开放性、多中心、随机研究。研究的主要终点指标是 OS。结果显示,对于 R/R DLBCL 患者,相较于对比利妥昔单抗联合 GemOx,格菲妥单抗联合 GemOx 的 OS 显著改善。在初步分析(中位随访期为 11.3 个月后进行)中,与利妥昔单抗联合 GemOx 相比,接受格菲妥单抗联合 GemOx 治疗的患者的死亡风险降低了 41%(风险比 [HR]=0.59,95% CI:0.40-0.89,p=0.011)。格菲妥单抗组合疗法还达到了其关键次要终点,与利妥昔单抗联合 GemOx 相比,PFS 降低了 63%(HR=0.37;95% CI:0.25–0.55,p<0.0001)。所有患者完成治疗后进行了随访分析(中位随访期为 20.7 个月),显示接受格菲妥单抗组合疗法治疗的患者中位 OS 为 25.5 个月,几乎是接受利妥昔单抗联合 GemOx 治疗的患者的两倍(vs 12.9 个月)。此外,获得完全缓解的患者数量是利妥昔单抗联合 GemOx 的两倍多(分别为 58.5% 和 25.3%)。罗氏在 CD20 靶点上已经有多款产品组合布局。除二代 CD20 单抗奥瑞利珠单抗外,罗氏在一代、三代中也均有代表产品,如一代的利妥昔单抗、三代的奥妥珠单抗,还有 2 款 CD3/CD20 双抗莫妥珠单抗和格菲妥单抗,另外 22 年 8 月还引进了一款 CD19/CD20 双靶点 CAR-T 疗法(RG6540)。3、卫材/渤健:仑卡奈单抗在欧盟获批上市当地时间 4 月 16 日,卫材与渤健共同宣布,欧盟委员会(EC)已经批准 Aβ 单抗 Leqembi® (lecanemab,仑卡奈单抗) 上市,治疗轻度认知障碍(MCI)以及由早期阿尔茨海默病(早期 AD)导致轻度痴呆的成年患者,这些患者为 ApoE ε4 非携带者或杂合子,且已确认存在淀粉样蛋白病变。该药因而成为首个在欧盟上市的同类药物。截图来自:企业官网EC 的批准主要基于 3 期临床试验 Clarify AD 的数据。Clarity AD 是一项全球验证性 III 期安慰剂对照、双盲、平行组、随机试验,在北美、欧洲和亚洲的 235 个研究中心纳入了 1,795 例早期 AD 患者,主要终点为 CDR-SB 评分。结果显示,在 EU 批准适应症对应人群(ApoE ε4 非携带者或杂合子)中,经过 18 个月的治疗,主要终点 CDR-SB 较基线的平均变化分别为 1.217 和 1.752 分,仑卡奈单抗较安慰剂显著降低体认知与功能量表评分 0.535 分(95% 置信区间 (CI):-0.778~ -0.293),降幅达 31%。相比安慰剂组,治疗组所有关键次要终点也显示出高度统计学显著差异。据 Insight 数据库显示,仑卡奈单抗此前已于 2023 年 1 月、2023 年 9 月、2024 年 1 月分别在美国、日本、中国获批上市。 重磅临床结果1、礼来:口服小分子 GLP-1RA 3 期临床结果积极,年内报上市当地时间 4 月 17 日,礼来公布了 3 期临床试验 ACHIEVE-1 的积极顶线结果,在该项研究中,每日一次 Orforglipron 相较于安慰剂在饮食和运动控制不佳的 2 型糖尿病患者中实现了积极结果。Orforglipron 由此成为首个 3 期临床成功的口服小分子 GLP-1R 激动剂。来源:企业官网在 ACHIEVE-1 研究(NCT05971940)中,Orforglipron 达到了主要终点,在 40 周时相较安慰剂实现了更佳的 A1C 减少,从基线 8% 降低了平均 1.3% 到 1.6%。关键次要终点而言,服用最高剂量 Orforglipron 的患者中,超过 65% 实现了 A1C ≤ 6.5%。减重次要终点上,最高剂量 Orforglipron 组患者平均减重 16.0 磅(7.9%,约 7.26 公斤)。具体疗效结果如下:礼来表示,将在后续的第 85 届 ADA 大会和学术期刊中发布 ACHIEVE-1 的详细结果。在今年晚些时候,还将公布 ACHIEVE 临床项目的更多结果,以及体重管理 3 期临床 ATTAIN 的数据。礼来预计将在 2025 年向全球监管机构递交 Orforglipron 用于体重管理的上市申请,在 2026 年度递交用于 2 型糖尿病的上市申请。 其他管线动态1、辉瑞再次放弃一款口服 GLP-1,肥胖管线仅剩 1 款药物当地时间 4 月 14 日,辉瑞宣布终止开发口服胰高血糖素样肽-1 (GLP-1) 受体激动剂 Danuglipron (PF-06882961)。截图来源:企业官网在接受每日一次 Danuglipron 治疗的 1400 多名患者中,肝酶升高的总体频率与同类已获批准的药物一致,但在其中一项剂量优化研究中,一名无症状参与者出现了潜在的药物性肝损伤,停用 Danuglipron 后病情得到缓解。在审查了所有信息后,包括迄今为止 Danuglipron 生成的所有临床数据和监管机构的最新意见,辉瑞决定停止该分子的开发。值得一提的是,在研究之初,辉瑞对于 Danuglipron 的设计是每日口服两次。2023 年 12 月,辉瑞公布该药治疗成人肥胖症的 IIb 期临床试验(NCT04707313)结果。该研究达到了主要终点,即体重较基线有显著变化。在安全性方面,最常见的不良事件为轻度且与机制相符的胃肠道反应,所有剂量组的停药率均高于 50%,而安慰剂组的停药率约为 40%。未报告新的安全信号,且与安慰剂组相比,Danuglipron 治疗未增加肝酶升高的发生率。基于该研究结果,辉瑞认为改良的每日一次 Danuglipron 有望在肥胖治疗领域发挥重要作用,后续将集中开发每日一次的制剂。然而此次出现的潜在肝损伤病例,每日一次的 Danuglipron 也终被辉瑞放弃。Danuglipron 并不是辉瑞在肥胖领域首次折戟的药物。此前 23 年 6 月,辉瑞宣布终止开发 Lotiglipron,这是一款每日一次口服的 GLP1R 激动剂。在先后终止 Lotiglipron 和 Danuglipron 开发后,目前辉瑞的肥胖药物管线仅剩一款口服 GIPR 拮抗剂PF-07976016,全球最高状态处于临床 II 期。 重磅医药交易据 Insight 数据库显示,本周(4 月 13 日 - 4 月 19 日)共发生 20 起交易事件。1、18.45 亿美元!赛诺菲重金加码自免和炎症性肠病领域当地时间 4 月 17 日,Earendil Labs 宣布与赛诺菲就两种潜在的首创双特异性抗体达成许可协议,用于自身免疫性和炎症性肠病领域。根据协议,赛诺菲将获得两种双特异性抗体 HXN-1002 和 HXN-1003 的全球独家授权,这两种抗体均将利用 Earendil Labs 专有的人工智能和高通量发现与研究平台进行开发。作为协议的一部分,Earendil Labs 将获得 1.25 亿美元的预付款,并有资格获得总计高达 17.2 亿美元的开发和商业里程碑付款,其中包括 5000 万美元的近期付款。此外,Earendil Labs 有资格获得从高个位数到低两位数不等的产品销售额的分级版税。HXN-1002 是一款靶向 α4β7 和 TL1A 的双特异性抗体。通过同时抑制两个经临床验证的靶点,HXN-1002 有望显著提高临床疗效,尤其适用于难治性患者。HXN-1002 旨在为中重度溃疡性结肠炎 (UC) 和克罗恩病 (CD) 患者提供一种治疗选择。HXN-1003 靶向 TL1A 和 IL23,这两个靶点是多种人类自身免疫性疾病中炎症反应的核心驱动因素。通过同时阻断这两条通路,该药已在结肠炎和皮肤炎症的临床前模型中展现出协同效应,有望解决尚未满足的治疗需求。2、3.57 亿美元!勃林格殷格翰重金加码自免赛道当地时间 4 月 15 日,勃林格殷格翰(BI)和 Cue Biopharma 宣布了一项战略研究合作和许可协议,以开发和商业化 Cue Biopharma 的 Cue -501 候选产品。根据协议条款,Cue Biopharma 的技术将被用于进一步研究和推进该候选分子的开发。双方未来还有可能扩大研究,并潜在开发针对多种自身免疫疾病的其他 B 细胞靶向双特异性药物。作为回报,Cue Biopharma 将获得 1200 万美元的预付款,此外,Cue Biopharma 还有资格获得总计约 3.45 亿美元的研发和商业里程碑付款(包括两个临床前开发里程碑)以及净销售额的特许权使用费。CUE-501 是一款 CD19/HLA 双抗,能够与 B 细胞特异性膜蛋白结合,同时选择性地与病毒特异性记忆杀伤性 T 细胞结合,使得 CUE-501 能够选择性地清除 B 细胞,抑制自身免疫和炎症反应。与其他靶向 B 细胞的疗法相比,CUE-501 有望提供更高的疗效和安全性,并有机会在患者治疗的早期阶段介入,实现对疾病的长期控制。3、最高 4.4 亿美元!橙帆医药首创双抗达成全球授权合作4 月 18 日,橙帆医药(VelaVigo)宣布,与美国 Ollin Biosciences 公司就自主研发的首创(FIC)双特异性抗体药物 VBS-102 达成全球独家授权协议。根据协议,Ollin 公司将获得该药物在大中华区以外的全球开发、生产及商业化权益,橙帆医药保留大中华区权益。 根据协议条款,本次交易总金额最高可达 4.4 亿美元,包含首付款、开发、注册及商业化里程碑付款。此外,橙帆医药将获得授权区域销售的分级特许权使用费。此次合作是橙帆医药在短短半年内达成的第二项国际授权协议,充分验证了公司创新研发平台的国际竞争力。目前,公司自主研发的首创(FIC)/同类最优(BIC)双抗 ADC 核心管线即将在美国进入临床阶段,另有五款 FIC/BIC 创新分子即将在全球行业会议上亮相。国内创新药进展本周国内共有 56 款创新药(含改良新)研发进度推进到了新阶段,其中 2 款获批上市,1 款申报上市,2 款首次启动 III 期临床,13 款获批临床,21 款申报临床。 新药获批上市1、康方生物:国内首个 IL-12/IL-23 单抗获批上市4 月 18 日,NMPA 显示,康方生物 1 类新药依若奇单抗在国内获批上市,适用于对环孢素、甲氨蝶呤(MTX)等其他系统性治疗或 PUVA(补骨脂素和紫外线 A)不应答、有禁忌或无法耐受的中度至重度斑块状银屑病的成年患者的治疗。截图来源:NMPA 官网依若奇单抗是国内首个针对 IL-12/IL-23 靶点的全新序列的 1 类抗体新药,可特异性、高亲和力结合至 IL-12/L-23 细胞因子两者共同的 p40 蛋白亚单位,通过抑制 Th1 和 Th17 细胞介导的异常免疫反应。依若奇单抗针对中重度斑块型银屑病患者共开展了 5 项临床研究,其中 2 项关键 III 期临床研究分别提供了依若奇单抗在中重度斑块型银屑病患者中的 16 周短期和 52 周长期的关键有效性和安全性数据。2023 年 EADV 年会上,康方展示了一项依若奇单抗 16 周短期用药的注册性随机安慰剂对照 III 期研究的结果(AK101-302)。结果显示,与安慰剂相比,接受依若奇单抗治疗的患者(135 mg 剂量给药)第 16 周 PASI75 应答率为 79.4%(vs 16.5%),sPGA0/1 达成率为 64%(vs 11.7%)。在安全性方面,依若奇单抗治疗中重度斑块型银屑病患者安全性良好,与同靶点药物相当;依若奇单抗用药产生抗药抗体(ADA)的发生率与同靶点药物一样低。2024 年 EADV 年会上,康方公布了依若奇单抗的一项注册性 III 期临床研究的 52 周长期疗效和安全性数据(AK101-303)。AK101-303 研究共入组 950 例中重度银屑病患者,包含了完成 AK101-302 研究且符合条件继续参加依若奇单抗长期治疗研究的患者(第 1 组和第 2 组)。研究结果显示,第 1 组患者在 AK101-302 研究中接受了依若奇单抗的 16 周治疗后,继续接受依若奇单抗治疗。这些患者在第 16 周时的 PASI75 应答率为 80.5%,sPGA0/1 达成率为 66.0%,并且这些改善在第 52 周仍然稳定维持。第 2 组患者在 AK101-302 研究中接受了对照药 16 周治疗后,第 16 周切换到依若奇单抗治疗,PASI75 应答率在第 32 周提升至 81.4%,sPGA0/1 达成率提升至 71.1%,并在第 52 周稳定维持。2、阿斯利康:国内首个 AKT 抑制剂获批上市4 月 18 日,NMPA 官网显示,阿斯利康 AKT 抑制剂卡匹色替片(Capivasertib)在国内获批上市,联合氟维司群用于转移性阶段至少接受过一种内分泌治疗后疾病进展,或在辅助治疗期间或完成辅助治疗后 12 个月内复发的激素受体(HR)阳性、人表皮生长因子受体 2(HER2)阴性且伴有一种或多种 PIK3CA/AKT1/PTEN 改变的局部晚期或转移性乳腺癌成人患者。值得一提的是,Capivasertib 是国内首个获批上市的 AKT 抑制剂。截图来自:NMPA 官网Capivasertib 是一种强效、选择性泛 AKT 抑制剂,于 2023 年 11 月首次在美国获批上市,用于治疗 HR 阳性、HER2 阴性乳腺癌。2022 年 SABCS 大会上,阿斯利康公布了一项全球多中心、双盲、随机的 III 期 CAPItello-291 临床试验结果。该试验旨在评估 capivasertib 联合氟维司群对比安慰剂联合氟维司群治疗局部晚期(不可手术)或转移性 HR 阳性、HER2 阴性乳腺癌的安全性和有效性。该研究共纳入 708 名受试者,41% 的受试者患有 PIK3CA、AKT1 或 PTEN 突变。研究结果显示,在所有受试者中,Capivasertib + 氟维司群和氟维司群 + 安慰剂组 ORR 分别为 22.9% vs 12.2%;mPFS 分别为 7.2 和 3.6 个月,与氟维司群+安慰剂组相比,Capivasertib + 氟维司群组疾病进展或死亡风险降低 40%。在 PIK3CA/AKT1/PTEN 信号通路变异亚组中,Capivasertib + 氟维司群和氟维司群+安慰剂组 ORR 分别为 28.8% vs 9.7%;mPFS 分别为 7.3 和 3.1 个月,与氟维司群+安慰剂组相比,Capivasertib + 氟维司群组疾病进展或死亡风险降低 50%。在安全性方面,Capivasertib 联合氟维司群组最常见的任何级别的不良事件(AEs)是腹泻、恶心、皮疹(包括皮疹、斑疹、斑丘疹和瘙痒性皮疹)、疲劳和呕吐。5% 及以上的患者最常发生的 3 级或以上的 AE 是腹泻和皮疹。除了乳腺癌,Capivasertib 还在拓展不同的适应症,包括前列腺癌(III 期)、非小细胞肺癌(II 期)和血液肿瘤(I 期)。 新适应症获批上市1、默沙东:九价 HPV 疫苗获批男性适应症4 月 14 日,默沙东九价人乳头瘤病毒疫苗 (酿酒酵母)-V503(商品名:佳达修 9)在国内获批新适应症,用于预防男性人乳头瘤病毒(HPV)感染(JXSS2300075/6)。截图来源:企业官网佳达修 9 是全球唯一一个获批的九价 HPV 疫苗,于 2014 年首次获 FDA 批准上市。2018 年 4 月,佳达修 9 首次在国内获批上市,目前其用药人群从最初的 16 - 26 岁女性拓展至 9 - 45 岁适龄女性。2024 年 1 月获 NMPA 批准新增二剂次接种程序适用于 9 - 14 岁女性。截图来源:Insight 数据库据 Insight 数据库显示,国内目前加入九价 HPV 疫苗赛道的有 7 家,其中万泰生物进展最快,正处于上市申请阶段。其他还有 4 家已进入 III 期临床,分别是康存惠、康乐卫士、瑞科生物和沃森生物。 申报上市1、诺诚健华:新一代泛 TRK 抑制剂在国内申报上市4 月 16 日,诺诚健华宣布,CDE 已受理其新一代泛 TRK 抑制剂卓乐替尼片(Zurletrectinib,ICP-723)的新药上市申请(NDA),用于治疗携带 NTRK 融合基因的晚期实体瘤成人和青少年(12 周岁≤年龄<18 周岁)患者。截图来源:CDE 官网NTRK 融合基因可见于各种类型的成人及儿童肿瘤。在部分罕见肿瘤中,例如唾液腺癌、分泌型乳腺癌、婴儿纤维肉瘤等,发生率超过 90%。中国新发的携带 NTRK 融合基因的肿瘤人群预估每年 6500 例,目前缺乏有效的治疗手段,存在未被满足的临床需求。在针对 NTRK 融合基因阳性的晚期实体瘤成人和青少年患者的关键注册临床试验中,卓乐替尼展示了卓越的有效性和安全性,同时可以克服第一代 TRK 抑制剂的耐药性。此外,卓乐替尼针对儿童患者(2 周岁≤年龄<12 周岁)的注册临床试验正在进行中,诺诚健华正加快临床研究,希望早日惠及儿童患者。2、礼来:IL-23 单抗新适应症在中国申报上市4 月 16 日,CDE 官网显示,礼来的 IL-23 单抗米吉珠单抗(Mirikizumab)在国内递交了一项新适应症上市申请。根据该药临床研究进展推测,本次申报的新适应症可能为溃疡性结肠炎。截图来源:CDE 官网米吉珠单抗可通过选择性靶向 IL-23 的 p19 亚基,阻断与 IL-23 受体的相互作用,抑制促炎细胞因子和趋化因子的释放,从而控制炎症。2023 年,该药相继在日本、欧盟、美国获批,用于治疗溃疡性结肠炎。2025 年以来,其又在欧美日获批治疗克罗恩病。在国内,米吉珠单抗在去年 10 月首次申报上市,并同时申报了静脉注射和皮下注射两种剂型,用于治疗克罗恩病。本次申报的新适应症推测为溃疡性结肠炎。米吉珠单抗溃疡性结肠炎适应症的 FDA 获批是基于 LUCENT 项目的结果,该研究旨在评估米吉珠单抗在中度至重度活动性溃疡性结肠炎 (UC) 成人患者中的疗效和安全性。 LUCENT 项目包括两项随机、双盲、安慰剂对照的Ⅲ期临床试验:一项是为期 12 周的诱导研究 (UC-1) ;另一项是为期 40 周的维持研究 (UC-2),共持续治疗 52 周。数据显示,在 UC-1 研究中,经过 12 周的治疗后:米吉珠单抗组有 24% 的患者获得临床缓解(vs 15%),65% 的患者获得临床反应 (vs 43%),34% 的患者获得内镜检查改善 (vs 21%),25% 的患者获得组织学内镜黏膜改善 (vs 14%),均高于安慰剂组。在 UC-2 研究中,接受米吉珠单抗治疗 40 周后,51% 的患者获得临床缓解,而安慰剂组为 27%。事后分析显示,在一年治疗后达到临床缓解的患者中,99% 的患者至少在之前的 12 周内未使用类固醇。此外,米吉珠单抗组有 50% 的患者达到不使用皮质类固醇的临床缓解 (vs 27%),58% 的患者内镜检查结果改善(vs 30%)。在第 12 周达到临床缓解的患者中,66% 的患者维持临床缓解(vs 40%)。43% 的患者组织学内镜黏膜改善 (vs 22%)。3、辉瑞:PARP 抑制剂「他拉唑帕利」新适应症国内申报上市4 月 16 日,CDE 官网显示,辉瑞「甲苯磺酸他拉唑帕利胶囊」新适应症上市申请获受理。根据注册分类和临床试验进展,Insight 数据库推测此次申报的适应症为胚系 BRCA 突变的 HER2 阴性乳腺癌。去年 11 月,该产品已在国内获批上市,联合恩扎卢胺用于 HRR 基因突变的转移性去势抵抗性前列腺癌 (mCRPC) 成人患者。来源:CDE 官网他拉唑帕利是一种聚腺苷二磷酸核糖聚合酶(PARP,包括 PARP1 和 PARP2)抑制剂,具有抑制 PARP 酶及 PARP 捕获双重作用机制。2018 年 10 月,他拉唑帕利首次在美国获批上市,用于治疗携带有害或疑似有害生殖细胞 BRCA 突变(gBRCAm)的 HER2 阴性局部晚期或转移性乳腺癌成年患者。2023 年 6 月,该产品新适应症获 FDA 批准,与恩扎卢胺联合用于 HRR 基因突变的 mCRPC 患者。 拟优先审评1、百济神州:BCL-2 抑制剂拟纳入优先审评4 月 17 日,CDE 官网显示,百济神州索托克拉片(Sonrotoclax)拟纳入优先审评,用于治疗既往接受过抗 CD20 治疗和 BTKi 治疗的套细胞淋巴瘤(MCL)成人患者和 CLL/SLL。根据 Insight 数据库,国内尚无 BCL-2 抑制剂获批治疗 MCL,索托克拉有望成为首个获批此适应症的 BCL-2 抑制剂。截图来源:CDE 官网索托克拉是百济神州开发的一款新一代 BCL-2 抑制剂。在国内,百济神州正在开展一项索托克拉 + 泽布替尼联合治疗复发/难治性套细胞淋巴瘤患者的 Ⅲ 期研究。在 2024 年财报中,百济神州曾表示,有望在今年下半年基于 Ⅱ 期研究数据递交索托克拉治疗 R/R CLL 和 R/R MCL 的加速批准申请。2024 年 EHA 会议上,百济神州曾公布了索托克拉联合泽布替尼治疗复发/难治性MCL 患者的Ⅰ期临床结果 (NCT04277637) 。该研究的主要终点是安全性,次要终点是 ORR。截至 2023 年 10 月 31 日,共入组 35 例 R/R MCL 患者,分为不同剂量组(80 mg,n=6;160 mg,n=12;320 mg,n=14;640 mg,n=3)。最大评估剂量 640 mg,未达到最大耐受剂量。无剂量限制性毒性 (DLT) 发生,研究者选择了 160 mg 和 320 mg 剂量水平的索托克拉进行扩展队列研究。在接受索托克拉 + 泽布替尼治疗的患者中,发生率≥20% 的 TEAE 包括中性粒细胞减少症、挫伤、血小板减少症和腹泻。中性粒细胞减少症是最常见的≥3 级 TEAE。未发生实验室或临床 TLS 病例,也未报告心房或心室颤动病例。27 例疗效可评估患者中,ORR 为 85%,其中 18 例完全缓解(CR;67%)。剂量扩展组中疗效可评估患者中,320 mg 组的 CR 率为 91%(10/11)、 ORR 为 91%(10/11);160 mg 组的 CR 率为 44%(4/9)、ORR 为 88%(8/9)。达到 CR 的中位时间为 6.4 个月。在 2 例既往 BTK 抑制剂治疗进展的疗效可评估患者中,观察到 1 例 CR 和 1 例 PD。2、鞍石生物:三代 EGFR TKI 拟纳入优先审评4 月 15 日,CDE 官网显示,北京鞍石生物科技股份有限公司旗下鞍石药业申报的苯甲酸安达替尼胶囊(PLB1004 胶囊)拟纳入优先审评,用于治疗既往经含铂化疗和/或 PD-1/PD-L1 免疫治疗时或治疗后出现疾病进展或不耐受,并且经检测确认存在 EGFR 20 号外显子插入突变的局部晚期或转移性 NSCLC 患者。截图来源:CDE 官网安达替尼是鞍石生物自主研发的新型小分子酪氨酸激酶不可逆抑制剂,具有高选择性,可以透过血脑屏障。非临床药效实验表明该化合物对 EGFR 20 号外显子插入(ex20 ins)突变、HER2 20 号外显子插入(ex20 ins)突变、EGFR 敏感突变、EGFR 耐药突变和 EGFR 罕见突变均有效。在 2023 年 AACR 大会上,鞍石生物公布安达替尼首次人体剂量递增和扩展研究(NCT05347628)的中期结果。这是一项评估安达替尼治疗晚期 NSCLC 的安全性、耐受性、药代动力学与抗肿瘤作用的 I 期多中心、开放、剂量递增与剂量扩展研究(PLB1004-I-01)。截至 2022 年 7 月 31 日,共 65 例受试者接受安达替尼治疗(剂量递增组 32 例,剂量扩展组 33 例)。截至 2022 年 7 月 31 日,≥160 mg QD 剂量水平共 29 例,其中 26 例受试者至少完成一次肿瘤评估,经研究者评估达到客观缓解的 15 例,ORR 为 57.7%;42.3%的受试者达到疾病稳定(SD),疾病控制率(DCR)为 100%。在 15 例 PR 受试者中,3 例受试者的持续缓解时间(DOR)已超过 240 天(其中 2 例受试者的 DOR 已超过 300 天,且仍在持续获益)。此外,亚组分析显示,在 ≥160 mg QD 剂量水平且至少完成一次肿瘤评估的 26 例携带 EGFR ex20ins 受试者中,8 例受试者基线时伴有脑转移,其中 3 例疗效评估达到 PR,ORR 达到 37.5%。接受安达替尼治疗的 65 例受试者均纳入本次安全性分析。剂量递增阶段的每个剂量组均无 DLT 发生,试验中未观察到 MTD。试验中不良事件大部分为 1~2 级。安达替尼在携带 EGFR ex20ins 的 NSCLC 患者中显示出较佳的疗效,无论患者是否有脑转移病灶;在安全性方面,试验中观察到的常见不良事件也在同类药物中均有所报道,经临床治疗后均可恢复,药物耐受性良好。Insight 数据库显示,鞍石生物针对该药已启动多个适应症研究,包括非鳞状非小细胞肺癌(临床 III 期)、非小细胞肺癌(临床 II 期)和非小细胞肺癌脑转移(临床 II 期)。 启动临床1、默沙东/科伦博泰:TROP2 ADC 启动第 12 项 III 期临床试验4 月 18 日,中国药物临床试验登记与信息公示平台显示,默沙东登记了一项比较 Sac-TMT(芦康沙妥珠单抗,TROP2 ADC)联合/不联合帕博利珠单抗与 TPC 一线治疗不可切除/转移性三阴性乳腺癌(PD-L1 CPS<10)的 III 期研究(CTR20251453)。截图来自:药物临床试验登记与信息公示平台这是一项 III 期、随机、开放标签研究(TroFuse-011),旨在既往未经系统治疗的局部复发性不可切除或转移性三阴性乳腺癌且 PD-L1 CPS<10 的受试者中比较 Sac-TMT 单药治疗、Sac-TMT 与帕博利珠单抗(MK-3475)联合治疗与医生选择的治疗的有效性和安全性。研究的主要终点是 PFS 和 OS。Insight 数据库显示,这是默沙东针对该药启动的第 12 项全球多中心临床试验。本次试验有 253 个机构参与,国内拟入组 144 人,国际入组 1000 人。此前还有 11 项临床试验,涉及卵巢癌、宫颈癌、胃癌、非小细胞肺癌等。Sac-TMT 已于 2024 年 11 月首次获 NMPA 批准上市,用于三阴性乳腺癌二线治疗。2025 年 3 月,该药在国内获批一项新适应症,用于治疗经 EGFR TKI 和含铂化疗治疗后进展的 EGFR 基因突变阳性的局部晚期或转移性非鳞状 NSCLC 成人患者。2、翰森制药:B7-H3 ADC 启动骨肉瘤 III 期临床4 月 18 日,中国药物临床试验登记与信息公示平台显示,翰森登记了一项注射用 HS-20093 对比吉西他滨联合多西他赛治疗既往二线治疗失败的骨肉瘤的 III 期临床研究(CTR20251474)。这也是该药启动的首个骨肉瘤 III 期临床。截图来自:药物临床试验登记与信息公示平台这是一项随机、对照、开放、多中心 III 期临床研究,旨在评估注射用 HS-20093 对比吉西他滨联合多西他赛治疗既往二线治疗失败的骨肉瘤有效性和安全性。国内拟入组 117 人。研究的主要终点是 PFS。HS-20093 是一款 B7-H3 ADC,采用经临床验证的拓扑异构酶抑制剂(TOPOi)有效载荷。2023 年 3 月,翰森曾启动一项该药治疗骨肉瘤的 II 期临床试验(ARTEMIS-002)。该结果首次在 2024 年 ASCO 大会上被公布。截至 2023 年 12 月 25 日,21 例可评估疗效患者(11 例接受 8 mg/kg 治疗,10 例接受 12.0 mg/kg 治疗)的中位随访时间为 4.1 个月:12.0 mg/kg HS-20093 剂量组的患者 ORR 为 20.0%。在 12.0 mg/kg 剂量组患者中观察到两例确认的部分缓解,并持续至末次随访,其中最长的缓解持续时间为 4.0 个月。 8 mg/kg 和 12.0 mg/kg 剂量组的疾病控制率分别为 81.8% 和 100%。21 例患者的中位无进展生存期数据均未达到成熟。在安全性方面,33 例患者发生治疗中出现的不良事件(TEAE)。常见的 3/4 级 TEAE(≥5%)为中性粒细胞减少、白细胞减少、血小板减少、淋巴细胞减少和贫血。无导致死亡的 TEAE。3、荣昌生物:维迪西妥单抗启动新 Ⅲ 期临床,HER2 低表达胃癌一线治疗4 月 16 日,药物临床试验登记与信息公示平台官网显示,荣昌生物登记了一项 III 期研究,以评估维迪西妥单抗联合替雷利珠单抗及 CAPOX 方案(奥沙利铂+卡培他滨)一线治疗 HER2 低表达的晚期胃/胃食管结合部腺癌的有效性和安全性,对照组为替雷利珠单抗联合 CAPOX。根据 Insight 数据库,全球尚无药物获批治疗 HER2 低表达胃癌,维迪西妥单抗在该适应症赛道进度属于第一梯队。截图来源:药物临床试验登记与信息公示平台官网维迪西妥单抗是一款靶向 HER2 的抗体偶联药物(ADC),此前已有两个适应症在国内获批上市,分别适用于至少接受过 2 种系统化疗的 HER2 过表达局部晚期或转移性胃癌患者、既往接受过含铂化疗且 HER2 过表达局部晚期或转移性尿路上皮癌患者,并被纳入国家医保目录。2024 年 10 月,维迪西妥单抗递交了第三项适应症的上市申请,并被 CDE 纳入优先审评,用于治疗 HER2 阳性存在肝转移的晚期乳腺癌患者,这些患者既往曾接受过曲妥珠单抗或其生物类似药和紫杉类药物治疗。本次启动的随机、对照 III 期研究适应症为一线治疗 HER2 低表达的晚期胃/胃食管结合部腺癌。该研究将在国内 80 家医疗机构开展,拟入组 616 例患者。研究的主要终点是 BIRC 根据 RECIST v1.1 评估的无进展生存期(PFS),次要终点指标包括生存期(OS)、研究者评估的 PFS 和客观缓解率 (ORR),以及疾病控制率(DCR)、缓解持续时间(DoR)等。目前,荣昌还在探索维迪西妥单抗治疗更多适应症的潜力。Insight 数据库显示,维迪西妥单抗有多项 Ⅲ 期临床正在进行中,除了本次新启动的 HER2 低表达胃癌一线,其它适应症还有尿路上皮癌/膀胱癌(HER2 表达|一线)、HER2 低表达乳腺癌(二线)、胃癌(HER2 阳性|三线)。4、恒瑞医药:CD79b ADC 启动首个 Ⅲ 期临床4 月 16 日,药物临床试验登记与信息公示平台官网显示,恒瑞医药启动了 CD79b ADC 产品 SHR-A1912 的国内 Ⅲ 期临床研究,以评估 SHR-A1912 联合利妥昔单抗+吉西他滨+奥沙利铂 (R-GemOx) 对比 R-GemOx 在复发/难治弥漫大 B 细胞淋巴瘤患者中的有效性和安全性。该 Ⅲ 研究将在国内 38 家医疗机构开展,拟入组 280 名患者。试验的主要终点是完全缓解率 (CRR) 和总生存期 (OS),次要终点是无进展生存期 (PFS)、不良事件的发生率及严重程度等。截图来源:Insight 数据库SHR-A1912 是恒瑞自主研发的一款以 CD79b 为靶点的 ADC,可以与表达 CD79b 的肿瘤细胞特异性结合,经肿瘤细胞内吞后在溶酶体内水解释放小分子毒素,从而诱导肿瘤细胞凋亡。2024 年 2 月,SHR-A1912 曾获得美国 FDA 授予快速通道资格,用于治疗既往接受过至少 2 线治疗的复发/难治性弥漫大 B 细胞淋巴瘤(R/R DLBCL)。2024 年 ASCO 会议上,恒瑞曾公布了 SHR-A1912 用于治疗 B 细胞非霍奇金淋巴瘤(B-NHL)患者的首次人体 Ⅰ 期临床试验结果。截至 2023 年 11 月 7 日,共入组 49 例患者。安全性数据显示,20 例(40.8%)患者发生了≥3 级治疗相关不良事件,其中最常见的(≥20%)是中性粒细胞计数下降。41 例基线时存在靶病变的患者至少进行了一次基线后评估,ORR 为 56.1%,6 个月 DoR 率为 62.7%。其中,DLBCL 患者的 ORR 为 51.9%(2.7 和 3.6 mg/kg 剂量组分别为 57.1% 和 77.8%),滤泡性淋巴瘤患者的 ORR 为 63.6%,边缘区淋巴瘤患者的 ORR 为 66.7%。总体而言,SHR-A1912 在高达 3.6 mg/kg 的剂量下仍具有耐受性,且在既往接受过治疗的 B-NHL 患者中表现出良好的抗肿瘤活性,Ⅰ 期结果支持对 SHR-A1912 进行进一步研究。5、信达生物:「玛仕度肽」启动首个 OSA 适应症 III 期临床4 月 14 日,药物临床试验登记与信息公示平台官网显示,信达生物登记了一项评估 IBI362(玛仕度肽)在患有中重度阻塞性睡眠呼吸暂停且肥胖的受试者中有效性和安全性的 III 期临床试验(GLORY-OSA)。这也是该药启动的首个中重度阻塞性睡眠呼吸暂停且肥胖的 III 期临床。截图来源:药物临床试验登记与信息公示平台官网这是一项随机、双盲、安慰剂对照的 III 期临床研究,旨在中国患有中重度阻塞性睡眠呼吸暂停且 BMI≥28kg/m2 的受试者中评估 IBI362 有效性和安全性。该研究计划国内入组 260 人。研究的主要终点是 48 周 AHI 自基线变化的幅度。玛仕度肽是信达生物与礼来共同开发的一款胰高血糖素样肽-1 受体(GLP-1R)/胰高血糖素受体(GCGR)双重激动剂。截至目前,玛仕度肽已经启动 7 项 III 期临床,涉及 2 型糖尿病、肥胖、MAFLD 和中重度阻塞性睡眠呼吸暂停且肥胖,其中 GLORY-1、DREAMS-1 和 DREAMS-2 研究均已达成终点。值得一提的是,该药当前共有 2 项 NDA(肥胖/2 型糖尿病)获 NMPA 受理审评中,预计本年陆续获批减重和 2 型糖尿病两项适应症。内容来源:药企官方发布新闻/资料、Insight 数据库封面来源:站酷海洛 Plus免责声明:本文仅作信息分享,不代表 Insight 立场和观点,也不作治疗方案推荐和介绍。如有需求,请咨询和联系正规医疗机构。PR 稿对接:微信 insightxb投稿:微信 insightxb;邮箱 insight@dxy.cn点击卡片进入 Insight 小程序国内审评进度、全球新药开发…随时随地查!多样化功能、可溯源数据……Insight 数据库网页版等你体验点击阅读原文,立刻解锁!
临床3期上市批准临床结果申请上市
2025-04-16
·医药观澜
▎药明康德内容团队报道今日(4月16日),中国国家药监局药品审评中心(CDE)官网最新公示,辉瑞(Pfizer)申报的甲苯磺酸他拉唑帕利胶囊新适应症上市申请获得受理,具体适应症尚未披露。公开资料显示,这是一款口服PARP抑制剂,此前于2024年11月首次获NMPA批准,与恩扎卢胺联合用于HRR(同源重组修复)基因突变的转移性去势抵抗性前列腺癌 (mCRPC) 成人患者。截图来源:CDE官网他拉唑帕利胶囊(talazoparib)是一种聚腺苷二磷酸核糖聚合酶(PARP,包括PARP1和PARP2)抑制剂,具有抑制PARP酶及PARP捕获双重作用机制。临床前研究表明,该产品可阻断PARP酶活性,并在DNA损伤部位捕获PARP,从而减少癌细胞生长,诱导癌细胞死亡。他拉唑帕利于2018年获得美国FDA批准(商品名为Talzenna),用于治疗患有有害或疑似有害生殖系乳腺癌易感基因(BRCA)突变(gBRCAm)人表皮生长因子受体2(HER2)阴性局部晚期或转移性乳腺癌的成人患者。2023年6月,FDA批准他拉唑帕利联合恩扎卢胺用于HRR基因突变的mCRPC成人患者的治疗。2024年1月,欧洲药品管理局(EMA)批准他拉唑帕利与恩扎卢胺联用治疗无临床化疗指征的mCRPC成人患者。根据中国药物临床试验登记与信息公示平台官网查询可知,他拉唑帕利的两项国际多中心(含中国)3期临床研究正在进行中,包括TALAPRO-3研究——他拉唑帕利与恩扎卢胺联合治疗DDR基因突变mCSPC男性患者;TALAPRO-2研究——他拉唑帕利和恩杂鲁胺联合治疗mCSPC患者。值得一提的是,2025年2月,辉瑞公布了TALAPRO-2临床3期研究的最新结果。这是一项多中心、随机、双盲、安慰剂对照研究,评估了两个mCRPC患者队列:队列1(所有患者,n=805)和队列2(HRR基因突变患者,n=399)。结果显示,与接受安慰剂联合恩扎卢胺治疗的患者相比,接受他拉唑帕利联合恩扎卢胺治疗的mCRPC患者,无论是否携带同源重组修复基因突变,总生存期(OS)显示出具有统计学显著和临床意义的改善。具体而言,经过超过四年的中位随访时间(52.5个月),与对照组恩扎卢胺单药相比,他拉唑帕利联合恩扎卢胺在全人群患者(队列1)中将中位总生存期(mOS)延长了近9个月,死亡风险降低了20%。队列2结果显示,在中位随访时间为44.2个月时,他拉唑帕利联合恩扎卢胺相较于对照组恩扎卢胺单药,将OS延长了14个月,死亡风险降低了38%。在携带HRR基因突变的患者群体中,无论是BRCA基因还是非BRCA基因突变的患者,均观察到了OS的改善。辉瑞此前新闻稿表示,TALAPRO-2的最新数据表明,无论是否有HRR基因突变,他拉唑帕利与恩扎卢胺联合用药均可显著延长mCRPC患者的OS,并有望重塑mCRPC治疗格局。参考资料:[1]中国国家药监局药品审评中心(CDE)官网. Retrieved Apr 16,2025, From https://www.cde.org.cn/main/xxgk/listpage/4b5255eb0a84820cef4ca3e8b6bbe20c[2]辉瑞公布最新研究数据:泰泽纳®联合恩扎卢胺可显著提升转移性去势抵抗性前列腺癌患者生存期并降低死亡风险. Retrieved Feb 14, 2025 from https://mp.weixin.qq.com/s/YShun3Ok0jhKtdgxBe8grg本文由药明康德内容团队根据公开资料整理编辑,欢迎个人转发至朋友圈。转发授权及其他合作需求,请联系wuxi_media@wuxiapptec.com。免责声明:药明康德内容团队专注介绍全球生物医药健康研究进展。本文仅作信息交流之目的,文中观点不代表药明康德立场,亦不代表药明康德支持或反对文中观点。本文也不是治疗方案推荐。如需获得治疗方案指导,请前往正规医院就诊。
临床3期申请上市上市批准
2025-03-02
·药时空
2025 年初,中国国家药监局药品审评中心(CDE)官网显示,多款 1 类创新药首次在中国获得临床试验默示许可(IND),这些新药来自多家知名药企,涵盖多种药物类型和适应症,展现了中国医药研发的活跃态势。
一、PF-07934040片
研发企业:辉瑞公司
作用机制:泛KRAS抑制剂
适应症:携带KRAS突变的晚期实体瘤
药品介绍:PF-07934040片是一款在研的泛KRAS抑制剂,正在国际范围内开展1期临床试验。该药物旨在开发出能够靶向多种、甚至全部KRAS突变体的泛KRAS(pan-KRAS)靶向疗法,为携带KRAS突变的诸多癌症类型提供治疗选择。二、XZP-7797-H1
研发企业:轩竹生物科技股份有限公司
作用机制:PARP1抑制剂
适应症:实体瘤
药品介绍:XZP-7797-H1是一款有效的、选择性的、能穿透血脑屏障的PARP1抑制剂。在生化分析中,该产品对PARP1的选择性超过PARP2和PARP家族其他成员的1000倍。临床前研究表明,该药物在BRCA1突变体乳腺癌异种移植模型中显示出剂量依赖性的快速肿瘤消退,在胰腺癌异种移植模型中也能显著抑制肿瘤生长。三、H1710注射用药品
研发企业:深圳市海普瑞药业集团股份有限公司
作用机制:乙酰肝素酶抑制剂
适应症:晚期实体瘤
药品介绍:H1710是一种乙酰肝素酶(HPA)抑制剂,能够有效抑制乙酰肝素酶活性,降低肿瘤细胞体外及肿瘤体内的乙酰肝素酶表达。临床前试验中,提示H1710在抑制肿瘤转移方面发挥效用;此外,单独应用H1710或联用多西他赛(DOX),能显著延长受治小鼠的存活时间。四、NKC007细胞注射液
研发企业:蒽恺赛生物
作用机制:非基因改造NK细胞产品
适应症:复发/难治性急性髓系白血病
药品介绍:NKC007细胞注射液是蒽恺赛生物自主研发的同种异体外周血来源的非基因改造NK细胞产品。蒽恺赛生物于2021年成立,是聚焦细胞与基因治疗的创新型生物医药公司,专注于开发针对癌症、感染性疾病和自身免疫性疾病的细胞治疗产品。五、NCR101注射液
研发企业:中盛溯源(广州)生物科技有限公司
作用机制:iPSC来源的基因修饰间充质样细胞(iMSC)
适应症:间质性肺病
药品介绍:NCR101是一款诱导多能干细胞(iPSC)来源的基因修饰间充质样细胞(iMSC)治疗产品。iPSC具备无限扩增的能力,可为现货型细胞治疗产品提供稳定且无限的种子细胞来源。NCR101作为iMSC产品,不仅能够实现规模化生产,还可以确保不同批次间iMSC的高度同质性。此外,该产品利用iPSC易于基因工程化改造的特性,通过基因修饰技术进一步增强了iMSC疗效。六、SIR2501片
研发企业:维泰瑞隆
作用机制:化药新药
适应症:肌萎缩侧索硬化(ALS)
药品介绍:SIR2501片的具体作用机制尚未公示。维泰瑞隆创立于2017年,创始人王晓东院士和张志远博士。该公司致力于开发能够从根本上解决衰老相关退行性疾病的变革性疗法,聚焦于衰老相关退行性疾病的致病机理,包括细胞程序性死亡,神经保护性通路、神经炎症等。七、JX10注射用药品
研发企业:箕星药业
作用机制:纤溶酶原调节剂
适应症:急性缺血性卒中
药品介绍:JX10(曾用名:BIIB131)是一款纤溶酶原调节剂,为具有抗炎特性的新型溶栓药物。该药的作用机制包括溶栓和抗炎,它可通过恢复急性中风后的临界血流量,延长原本短暂的治疗窗口期,即发病后4.5小时内,使更多患者可以从目前的标准治疗中获益。八、MK-6070注射用药品
研发企业:默沙东(MSD)和第一三共(Daiichi Sankyo)
作用机制:靶向DLL3的三特异性T细胞接合器
适应症:小细胞肺癌
药品介绍:MK-6070(曾用名:HPN328)是一种靶向DLL3的三特异性T细胞接合器,目前默沙东和第一三共正在全球范围内开发该产品。该产品包含3个结合域,用于引导T细胞杀死表达DLL3的癌细胞。九、deucrictibant缓释片
研发企业:Pharvaris公司
作用机制:缓激肽B2受体拮抗剂
适应症:遗传性血管性水肿
药品介绍:deucrictibant缓释片是缓激肽B2受体的一种强效、选择性的口服拮抗剂,目前正在国际范围内开展3期临床研究。在遗传性血管性水肿(HAE)中,缓激肽B2受体的拮抗作用是一种耐受性良好、已被临床证实的作用机制。通过抑制缓激肽信号传导,deucrictibant有可能治疗HAE发作的临床体征并预防发作的发生。十、HB0056注射液
研发企业:华奥泰生物和华博生物
作用机制:靶向TSLP和IL-11双抗
适应症:中重度哮喘
药品介绍:HB0056是一款同时靶向TSLP和IL-11的双特异性抗体,它能同时阻断IL-11与TSLP信号通路,抑制其生物学活性。同时,该产品可抑制MRC-5细胞(人胚肺成纤维细胞)纤维化相关基因的表达,并在研究过程中体现出双靶点协同增效的作用潜力。十一、JYP0015片
研发企业:嘉越医药
作用机制:泛RAS抑制剂
适应症:RAS突变的实体瘤和血液瘤
药品介绍:JYP0015是一款泛RAS抑制剂,为一款分子胶产品,它可以通过形成RAS-亲环蛋白A蛋白复合物来阻止RAS介导的信号传导。2024年5月,嘉越医药已经与ERASCA公司达成一项全球独家授权协议,将JYP0015在中国内地和香港、澳门地区之外的全球独家研究、开发和商业化权利授予后者。十二、INV-6452片
研发企业:广东省创新药转化医学研究院有限公司和原力生命科学有限公司
作用机制:靶向CDK2/4的化药新药
适应症:乳腺癌或实体瘤
药品介绍:INV-6452片靶向CDK2/4,拟开发治疗激素受体阳性、人表皮生长因子受体2阴性的(HR+/HER2-)晚期/转移性乳腺癌或局部晚期/转移性实体瘤。十三、GC310腺相关病毒注射液
研发企业:锦篮基因
作用机制:AAV基因治疗药物
适应症:肝豆状核变性
药品介绍:GC310腺相关病毒注射液是锦篮基因自主研发的一款用于治疗肝豆状核变性的AAV基因治疗药物。非临床研究显示出良好的药物安全性和显著的疗效,单次治疗后的靶组织可表达具有生物学功能的miniATP7B铜离子转运蛋白,恢复铜离子代谢能力并提升铜蓝蛋白水平,有望长期改善肝豆状核变性患者的现状。十四、ICG318 CAR-T细胞注射液
研发企业:松鹤免疫生物
作用机制:CAR-T细胞疗法
适应症:系统性红斑狼疮
药品介绍:ICG318 CAR-T细胞注射液拟开发治疗成人难治性系统性红斑狼疮。CAR-T细胞疗法在癌症治疗之外,正在自身免疫疾病发挥治疗潜力,系统性红斑狼疮是正在广泛被探索的适应症之一。十五、HPB-143片
研发企业:多域生物
作用机制:靶向IRAK4的蛋白降解靶向嵌合体
适应症:特应性皮炎等自身免疫性和炎症性疾病
药品介绍:HPB-143片是一款靶向IRAK4的蛋白降解靶向嵌合体(简称PROTAC)。IRAK4蛋白同时具有激酶活性以及支架功能,因此利用降解剂对其进行降解能同时阻断这两个功能,从而实现对相关信号通路的更全面的抑制,充分发挥抗炎活性。十六、AWT020注射用无菌粉末
研发企业:君实生物
作用机制:PD-1和IL-2双功能性抗体融合蛋白
适应症:晚期恶性肿瘤
药品介绍:AWT020注射用无菌粉末是PD-1和白细胞介素-2(IL-2)双功能性抗体融合蛋白,主要用于晚期恶性肿瘤的治疗。鉴于PD-1和IL-2在肿瘤微环境中的共表达,该融合蛋白可在阻断PD-1通路的同时,通过与IL-2受体结合选择性地激活IL-2信号通路,从而增强抗肿瘤免疫反应。十七、AA001单抗
研发企业:智源鸿晟生物
作用机制:单抗药物
适应症:阿尔茨海默病(AD)源性轻度认知障碍及轻中度AD
药品介绍:AA001是一款治疗用单克隆抗体药物。智源生物刘瑞田教授研发团队通过十余年的探索,发现了阿尔茨海默病抗体药物介导神经突触过度丢失是导致免疫治疗失败的原因,提出采用无效应片段(Fc段)或无效应功能的β-淀粉样蛋白(Aβ)抗体具有较好前景的AD治疗新策略,可以提高抗体药物的疗效,降低毒副作用。十八、ZHB601注射用药品
研发企业:众红生物
作用机制:化药新药
适应症:脑小血管病
药品介绍:ZHB601注射用药品拟开发治疗脑小血管病。脑小血管病(cSVD)是指各种病因影响脑内小动脉、微动脉、毛细血管、微静脉和小静脉所致的一系列临床、影像和病理综合征。腔隙性缺血性卒中(“脑梗”的主要类型之一)是脑小血管病的主要临床表现之一。十九、GLS-W1100胶囊
研发企业:远大生物制药
作用机制:生物制品新药
适应症:外阴阴道假丝酵母菌病
药品介绍:GLS-W1100胶囊拟用于外阴阴道假丝酵母菌病的治疗。外阴阴道假丝酵母菌病(VVC)是临床常见的生殖道炎症,造成患者外阴瘙痒、分泌物增多等不适,白假丝酵母菌是其最常见的致病原。二十、F1F3软膏
研发企业:中奥生物医药
作用机制:外用多肽软膏
适应症:HPV引起的尖锐湿疣
药品介绍:F1F3软膏是中奥生物医药自主研发的一款外用多肽软膏。该产品具有双重作用机制:通过诱导HPV+肿瘤细胞焦亡并分泌促炎细胞因子,如白介素18,激活非特异性免疫反应;随后使T细胞浸润,激活特异性免疫反应,有助于预防尖锐湿疣的复发。二十一、KNT-0916片
研发企业:科恩泰生物医药科
作用机制:FGFR2高选择抑制剂
适应症:FGFR2异常实体瘤
药品介绍:KNT-0916为一款FGFR2高选择抑制剂。成纤维细胞生长因子受体(FGFR)的基因变异存在于多种类型的人类肿瘤中,FGFR信号通过促进肿瘤细胞增殖、存活、迁移和血管生成来促进恶性肿瘤的发展。二十二、IBI3017注射用药品
研发企业:信达生物
作用机制:生物制品新药
适应症:实体瘤
药品介绍:IBI3017拟用于治疗不可切除的局部晚期或转移性实体瘤。目前尚未从公开资料查询到该产品的作用机制,根据受理号可知,这是一款生物制品新药。二十三、DC50292A片
研发企业:双鹤润创
作用机制:化药新药
适应症:MTAP缺失实体瘤
药品介绍:DC50292A片拟开发治疗甲硫腺苷磷酸化酶(MTAP)缺失的晚期或转移性实体瘤。目前尚未从公开渠道查询到该产品的作用机制,从受理号可知这是一款化药新药。研究表明,MTAP基因缺失发生在大约10%的癌症中,包括胰腺癌、肺癌和膀胱癌等,它通常与PRMT5抑制剂构成“合成致死”效应。二十四、IBI3002注射用药品
研发企业:信达生物制药(苏州)有限公司
作用机制:抗IL-4Rα/TSLP双特异性抗体
适应症:哮喘、特应性皮炎
药品介绍:IBI3002是信达生物自主研发的一款抗IL-4Rα/TSLP双特异性抗体,目前正在澳大利亚开展1期临床研究。IBI3002具有高效的IL-4Rα和TSLP共同阻断功能,具有抑制2型和非2型炎症的潜力,在抑制2型炎症方面具有潜在的协同作用。二十五、HRS-1301片
研发企业:山东盛迪医药有限公司
作用机制:1类化学药物
适应症:高脂血症
药品介绍:HRS-1301片是恒瑞医药研发的1类新药,拟用于高脂血症治疗。具体作用机制尚未披露,临床前研究显示,该产品可有效改善高脂血症。目前全球范围内暂无同类药物获批上市。二十六、SLN12140注射用药品
研发企业:领诺(上海)医药科技有限公司
作用机制:单域抗体补体靶向药物
适应症:原发性IgA肾病
药品介绍:SLN12140是一款新一代补体靶向药物,基于单域抗体VHH-Fc结构形式,能够有效抑制IgAN病理过程中的补体异常活化。与在研的其他补体靶向药物(小分子或重组抗体)相比,其既有重组抗体较为稳定的药物动力学特征、较好的靶点特异性和药物活性,又有小分子药物给药方便(皮下给药)的特征。二十七、HS-10501-2注射用药品
研发企业:翰森制药
作用机制:化药新药
适应症:成人肥胖症和成人2型糖尿病
药品介绍:HS-10501-2拟开发治疗成人肥胖症和成人2型糖尿病。从受理号可知这是一款化药新药,目前尚未从公开渠道查询到该产品的具体作用机制。翰森制药另一款在研的口服GLP-1R激动剂药物HS-10501片已经在中国进入临床研究阶段,拟开发治疗成人肥胖症和2型糖尿病。二十八、Tye1001注射用药品
研发企业:泰尔康生物
作用机制:抗肿瘤偶联药物
适应症:晚期实体瘤和淋巴瘤
药品介绍:Tye1001是泰尔康生物自主研发的广谱抗肿瘤药物,是由高活高毒的毒素小分子与特定药物载体偶联的抗肿瘤偶联药物,主要通过肿瘤血管的EPR效应及细胞表面特异性受体靶向作用于实体瘤,通过细胞内吞方式进入肿瘤细胞,再经溶酶体降解、释放毒素小分子,毒素小分子靶向作用于细胞微管,抑制肿瘤生长。二十九、CXJM-66注射液
研发企业:人福药业
作用机制:钠离子通道阻断剂
适应症:手术麻醉;急性疼痛控制
药品介绍:CXJM-66是人福药业自主开发的新分子实体药物,为一种结构新颖的钠离子通道阻断剂,能阻断钠离子流入细胞膜内,抑制神经元兴奋信号的传导且不易洗脱,从而产生持久麻醉作用。非临床研究表明,CXJM-66注射液单次给药即可显著发挥镇痛作用,且未观察到明显毒性反应,安全性良好。三十、DXC008注射用药品
研发企业:多禧生物
作用机制:靶向STEAP1和PSMA的ADC
适应症:前列腺癌等多种实体瘤
药品介绍:DXC008是一种抗体偶联药物(ADC),由一种新型STEAP1抗体与管溶素类似物缀合物连接,它不仅对STEAP1具有良好的亲和性,而且对PSMA也具有中等的亲和性,有潜力治疗前列腺癌等肿瘤。STEAP1是一种在前列腺癌中频繁表达的细胞表面蛋白,在非前列腺组织中表达有限。三十一、CS12088片
研发企业:微芯生物
作用机制:HBV核衣壳组装调节剂
适应症:慢性乙肝
药品介绍:CS12088属于HBV核衣壳组装调节剂。该产品通过干扰HBV核衣壳的组装,阻断病毒前基因组RNA包装入核衣壳,进而阻断HBV DNA的复制,抑制成熟病毒颗粒的产生,且CS12088对HBV不同基因型均能保持纳摩尔级别的抗病毒活性。
识别微信二维码,可添加药时空小编
请注明:姓名+研究方向!
临床申请临床1期细胞疗法
分析
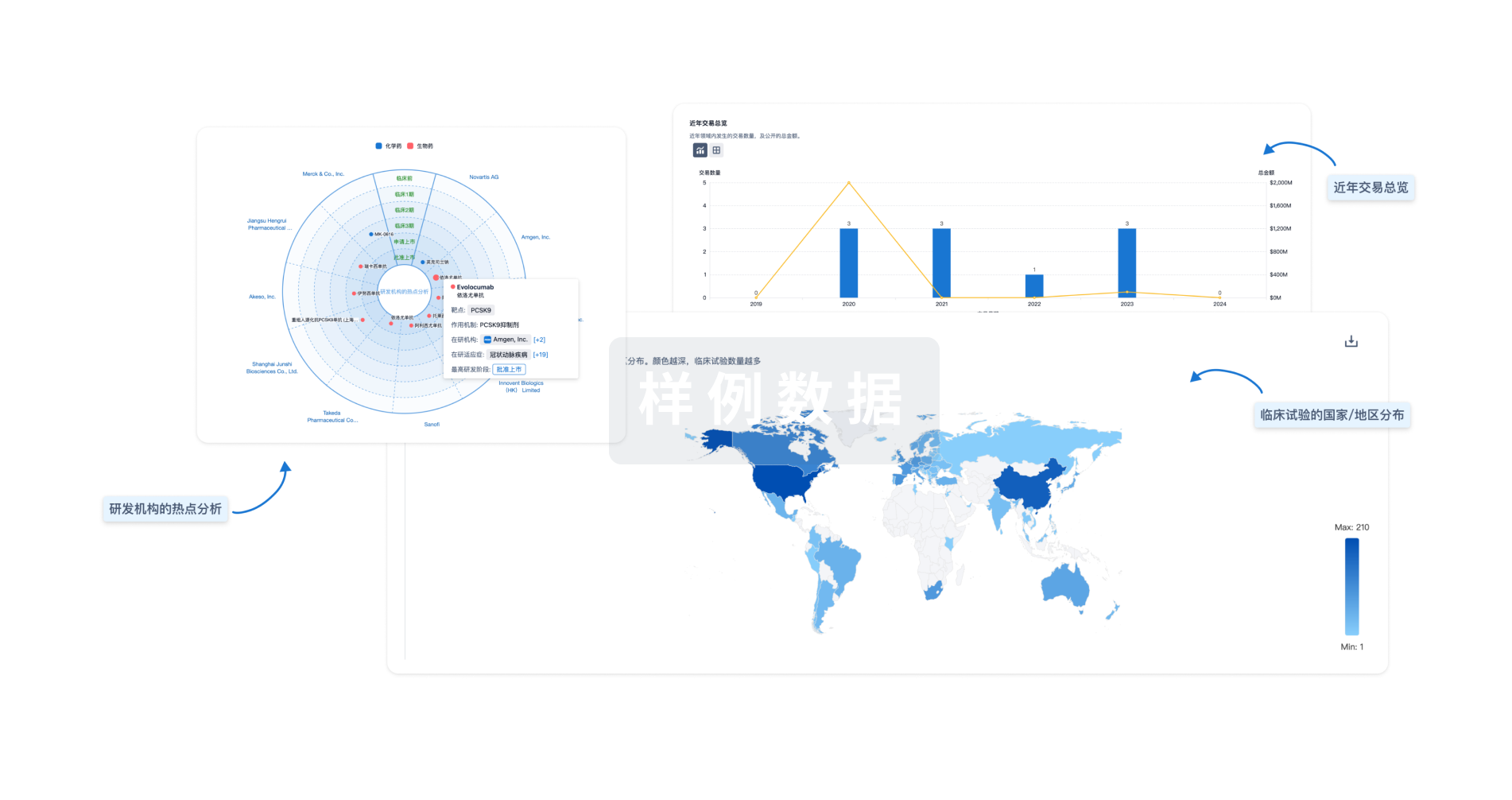
对领域进行一次全面的分析。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用




