预约演示
更新于:2025-05-07
RARγ
更新于:2025-05-07
基本信息
别名 NR1B3、Nuclear receptor subfamily 1 group B member 3、RAR-gamma + [5] |
简介 Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence of ligand, acts mainly as an activator of gene expression due to weak binding to corepressors. Required for limb bud development. In concert with RARA or RARB, required for skeletal growth, matrix homeostasis and growth plate function (By similarity). |
关联
31
项与 RARγ 相关的药物作用机制 RARα激动剂 [+2] |
在研适应症 |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2021-07-26 |
靶点 |
作用机制 RARγ激动剂 |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2019-10-04 |
175
项与 RARγ 相关的临床试验NCT06908954
A Phase I, Open-label, Parallel-group Study to Evaluate the Single-dose Pharmacokinetics of Palovarotene in Male and Female Participants With Moderate and Severe Hepatic Impairment and Matched Participants With Normal Hepatic Function
The main aim of this study is to understand how moderate and severe liver impairment (based on the Child-Pugh classification) affects the body's processing of a single dose of 10 mg maximum of palovarotene, compared to healthy participants with normal liver function.
The study will also assess the safety and tolerability of the single dose of palovarotene.
Participants will be enrolled in stages and divided into three groups based on their liver function:
* Group 1: Healthy participants with normal liver function
* Group 2: Participants with moderate liver impairment
* Group 3: Participants with severe liver impairment (only enrolled if Group 2 results are safe and acceptable)
Blood samples will be taken to assess how the drug binds to proteins in the blood. Participants will undergo various safety checks and procedures. Participants will stay in the clinical unit until Day 5 for these assessments and will return on Day 10 for a final visit.
The study will also assess the safety and tolerability of the single dose of palovarotene.
Participants will be enrolled in stages and divided into three groups based on their liver function:
* Group 1: Healthy participants with normal liver function
* Group 2: Participants with moderate liver impairment
* Group 3: Participants with severe liver impairment (only enrolled if Group 2 results are safe and acceptable)
Blood samples will be taken to assess how the drug binds to proteins in the blood. Participants will undergo various safety checks and procedures. Participants will stay in the clinical unit until Day 5 for these assessments and will return on Day 10 for a final visit.
开始日期2025-03-14 |
申办/合作机构 |
CTRI/2024/08/072412
A prospective, multi center, open label phase IV clinical study to evaluate the safety and efficacy of Trifarotene or Aklief 50 microgram per gram cream in subjects with acne vulgaris over 12 weeks. - NIL
开始日期2024-08-26 |
申办/合作机构 |
NCT06733402
A Multi-center, Double-blind, Randomized, Placebo Controlled, Parallel-group Study, Comparing Trifarotene Cream 0.005% (Taro Pharmaceuticals U.S.A, Inc.) to Aklief® Cream (US REFERENCE LISTED DRUG), Aklief Cream (REFERENCE PRODUCT) and Each Active Treatment to a Placebo Control in the Treatment of Acne Vulgaris.
To demonstrate the efficacy, therapeutic equivalence and safety of Trifarotene Cream 0.005% (Taro Pharmaceuticals U.S.A., Inc.) and US Reference Listed Drug, Canadian Reference Product over the placebo control in the treatment of acne vulgaris.
开始日期2024-06-20 |
100 项与 RARγ 相关的临床结果
登录后查看更多信息
100 项与 RARγ 相关的转化医学
登录后查看更多信息
0 项与 RARγ 相关的专利(医药)
登录后查看更多信息
1,191
项与 RARγ 相关的文献(医药)2025-07-01·Critical Reviews in Oncology/Hematology
Pharmacogenomics in pediatric oncology patients with solid tumors related to chemotherapy-induced toxicity: A systematic review
Review
作者: Uvdal, Hanna ; Hansson, Paula ; Blacker, Christopher ; Wadelius, Mia ; Green, Henrik ; Ljungman, Gustaf
2025-04-21·Journal of Bone and Mineral Research
Retinoid-impregnated nanoparticles enable control of bone growth by site-specific modulation of endochondral ossification in mice
Article
作者: Suzuki, Akiko ; Alferiev, Ivan ; Tian, Hongying ; Matsuoka, Masatake ; Abzug, Joshua M ; Oichi, Takeshi ; Iwamoto, Masahiro ; Enomoto-Iwamoto, Motomi ; Otsuru, Satoru ; Pacifici, Maurizio ; Herzenberg, John E ; Tang, Ningfeng ; Usami, Yu ; Uchibe, Kenta ; Chorny, Michael
2025-04-01·Cancer Genetics
Complex genetic structural aberrations revealed by optical genome mapping in a case of APL-like morphology
Article
作者: Karrs, Jeremiah X ; Steinmetz, Heather B ; Smuliac, Narcisa A ; Murad, Farzana ; Sathyanarayana, Shivaprasad H ; Sullivan, Matthew R ; Bickford, Michelle A ; Khan, Wahab A ; Bao, Jing ; Kaur, Prabhjot ; Tonseth, Kyle A
21
项与 RARγ 相关的新闻(医药)2024-12-01
11月28日,复旦大学研究团队在期刊《Advanced Science》上发表了题为“Synthetic Retinoid Sulfarotene Selectively Inhibits Tumor-Repopulating Cells of Intrahepatic Cholangiocarcinoma via Disrupting Cytoskeleton by P-Selectin/PSGL1 N-Glycosylation Blockage”的研究论文,本研究中,研究人员发现尽管5-氟尿嘧啶、顺铂、佩米加汀和吉西他滨都能抑制ICC-TRCs,但硫福罗汀的效能更为显著。硫福罗汀能将视黄醇受体α(RARɑ)从细胞质转运至细胞核,并通过转录水平抑制P-选择素的表达。此外,它还能直接与FUT8相互作用。这些作用共同作用,通过破坏PSGL1-调控的细胞骨架来抑制ICC-TRCs。这些发现为通过破坏P-选择素/PSGL1相互作用和改变PSGL1糖基化模式来破坏细胞骨架完整性和消除ICC-TRCs提供了策略。
11月28日,复旦大学研究团队在期刊《Advanced Science》上发表了题为“Synthetic Retinoid Sulfarotene Selectively Inhibits Tumor-Repopulating Cells of Intrahepatic Cholangiocarcinoma via Disrupting Cytoskeleton by P-Selectin/PSGL1 N-Glycosylation Blockage”的研究论文,本研究中,研究人员发现尽管5-氟尿嘧啶、顺铂、佩米加汀和吉西他滨都能抑制ICC-TRCs,但硫福罗汀的效能更为显著。硫福罗汀能将视黄醇受体α(RARɑ)从细胞质转运至细胞核,并通过转录水平抑制P-选择素的表达。此外,它还能直接与FUT8相互作用。这些作用共同作用,通过破坏PSGL1-调控的细胞骨架来抑制ICC-TRCs。
这些发现为通过破坏P-选择素/PSGL1相互作用和改变PSGL1糖基化模式来破坏细胞骨架完整性和消除ICC-TRCs提供了策略。
https://onlinelibrary-wiley-com.libproxy1.nus.edu.sg/doi/full/10.1002/advs.202407519
https://onlinelibrary-wiley-com.libproxy1.nus.edu.sg/doi/full/10.1002/advs.202407519
背景信息
背景信息
01
01
肝内胆管癌(ICC)在中国和东南亚地区发病率尤其高,是仅次于肝细胞癌的原发性肝癌第二大类型,占肝恶性肿瘤的20%左右,占所有消化系统恶性肿瘤的3%。这是一种高度侵袭性和致命的疾病,5年总体生存率(OS)仍维持在9%左右。仅有20%-30%的患者在可切除阶段被诊断出来,但即使经过手术切除,5年生存率也只有约20%-35%。相比之下,大多数不可切除患者的中位生存期(mOS)为11.7个月。尽管手术和系统性治疗的领域正在迅速发展,但治疗效果仍然不佳。因此,迫切需要探索更有效的ICC治疗方法。
肝内胆管癌(ICC)在中国和东南亚地区发病率尤其高,是仅次于肝细胞癌的原发性肝癌第二大类型,占肝恶性肿瘤的20%左右,占所有消化系统恶性肿瘤的3%。这是一种高度侵袭性和致命的疾病,5年总体生存率(OS)仍维持在9%左右。仅有20%-30%的患者在可切除阶段被诊断出来,但即使经过手术切除,5年生存率也只有约20%-35%。相比之下,大多数不可切除患者的中位生存期(mOS)为11.7个月。尽管手术和系统性治疗的领域正在迅速发展,但治疗效果仍然不佳。因此,迫切需要探索更有效的ICC治疗方法。
肿瘤再生细胞(TRCs)代表了一群自我更新能力强、高度肿瘤形成的癌细胞亚群,在癌症的关键事件中发挥着关键作用。这些事件包括维持细胞增殖、抵抗细胞死亡、远距离转移、免疫逃避、代谢重编程、衰老相关分泌表型以及耐药性等。为了消除这些TRCs,本研究小组开发了一种新型合成维甲酸,称为硫福罗汀(WYC-209,SFT)。在体外和体内评估,包括患者来源的异种移植(PDX)模型中,SFT对各种癌症类型的TRCs显示出卓越的抑制作用,因此它被用作潜在的TRCs靶向分子。SFT对TRCs的选择性抑制可能有助于解决上述ICC问题。
肿瘤再生细胞(TRCs)代表了一群自我更新能力强、高度肿瘤形成的癌细胞亚群,在癌症的关键事件中发挥着关键作用。这些事件包括维持细胞增殖、抵抗细胞死亡、远距离转移、免疫逃避、代谢重编程、衰老相关分泌表型以及耐药性等。为了消除这些TRCs,本研究小组开发了一种新型合成维甲酸,称为硫福罗汀(WYC-209,SFT)。在体外和体内评估,包括患者来源的异种移植(PDX)模型中,SFT对各种癌症类型的TRCs显示出卓越的抑制作用,因此它被用作潜在的TRCs靶向分子。SFT对TRCs的选择性抑制可能有助于解决上述ICC问题。
FUT8是ICC-TRCs中SFT的直接靶点
FUT8是ICC-TRCs中SFT的直接靶点
02
02
糖基化是在细胞内质网/高尔基体中的一种酶促过程,是蛋白质翻译后修饰的主要方式之一,由不同的糖基转移酶和/或糖苷酶催化。鉴于SFT对RARɑ的影响,研究人员首先推测它可能通过增加核内RARɑ的转运来调节糖基转移酶/糖苷酶的转录。经过交集运算后,研究人员重点关注了MGAT3基因,它是N-乙酰葡萄糖胺转移酶III的编码基因,通过β1,4糖苷键将N-乙酰葡萄糖胺连接到β-岩藻糖上,从而催化N-糖链核心的三糖N-甘露糖的β1,4糖苷键的形成。RNA-seq和qRT-PCR数据表明,SFT处理显著降低了ICC-TRCs中MGAT3 mRNA的水平,但在2D ICC细胞中未见明显变化。因此,研究人员使用siRNA沉默ICC-TRCs中的MGAT3。然而,沉默MGAT3未能抑制PSGL1的糖基化、细胞球体的生长、迁移和侵袭。这些结果表明,MGAT3的变化只是SFT治疗后的附加效应,而不是SFT抑制ICC-TRCs的原因。
糖基化是在细胞内质网/高尔基体中的一种酶促过程,是蛋白质翻译后修饰的主要方式之一,由不同的糖基转移酶和/或糖苷酶催化。鉴于SFT对RARɑ的影响,研究人员首先推测它可能通过增加核内RARɑ的转运来调节糖基转移酶/糖苷酶的转录。经过交集运算后,研究人员重点关注了MGAT3基因,它是N-乙酰葡萄糖胺转移酶III的编码基因,通过β1,4糖苷键将N-乙酰葡萄糖胺连接到β-岩藻糖上,从而催化N-糖链核心的三糖N-甘露糖的β1,4糖苷键的形成。RNA-seq和qRT-PCR数据表明,SFT处理显著降低了ICC-TRCs中MGAT3 mRNA的水平,但在2D ICC细胞中未见明显变化。因此,研究人员使用siRNA沉默ICC-TRCs中的MGAT3。然而,沉默MGAT3未能抑制PSGL1的糖基化、细胞球体的生长、迁移和侵袭。这些结果表明,MGAT3的变化只是SFT治疗后的附加效应,而不是SFT抑制ICC-TRCs的原因。
研究人员从HUCCT1-TRCs中提取总蛋白,并用DMSO或SFT(1 mm)处理。经胰蛋白酶消化后,使用质谱进行分析。生物信息学分析确定这些4047个肽段代表了1791种不同的蛋白质。随后,使用数据库将蛋白质转换为对应的编码基因。将它们与糖基转移酶或糖苷酶编码基因进行交集,总共获得了10个潜在的直接靶点,包括OGT、MGAT2、GALNT2、GALNT6、GALNT7、PIGT、FUT8、POFUT1、ALG2和UGGT1。RNA-seq数据表明,SFT治疗后这些基因的表达没有显著变化。研究人员推测,SFT可能通过直接与它们结合来调节酶的活性。
研究人员从HUCCT1-TRCs中提取总蛋白,并用DMSO或SFT(1 mm)处理。经胰蛋白酶消化后,使用质谱进行分析。生物信息学分析确定这些4047个肽段代表了1791种不同的蛋白质。随后,使用数据库将蛋白质转换为对应的编码基因。将它们与糖基转移酶或糖苷酶编码基因进行交集,总共获得了10个潜在的直接靶点,包括OGT、MGAT2、GALNT2、GALNT6、GALNT7、PIGT、FUT8、POFUT1、ALG2和UGGT1。RNA-seq数据表明,SFT治疗后这些基因的表达没有显著变化。研究人员推测,SFT可能通过直接与它们结合来调节酶的活性。
硫福罗汀直接与FUT8结合
硫福罗汀直接与FUT8结合
为了确定直接靶点,研究人员评估了ICC中10个潜在靶点的重要性。差异分析显示,OGT、GALNT2、GALNT6、GALNT7、PIGT、FUT8、POFUT1、ALG2和UGGT1在ICC组织中的表达水平高于正常组织。生存分析显示,只有FUT8与患者的总生存期(OS)和无复发生存期(RFS)显著相关。SFT治疗不会对FUT8的mRNA或蛋白水平产生显著影响。研究人员通过将这10个残基替换为ALA构建了突变型FUT8,并纯化了FUT8WT和FUT8MT蛋白。MST数据表明,SFT与FUT8WT蛋白具有很强的亲和力,但与FUT8MT蛋白的亲和力降低,Kd值分别为9.83E-08和2.83E-07。模拟对接数据表明,SFT与SER245、ASN247、GLY253、THR267形成稳定的氢键相互作用,与ILE242、TRP255、PRO261形成疏水相互作用。因此,SFT可能直接与ICC-TRC中的FUT8结合。
为了确定直接靶点,研究人员评估了ICC中10个潜在靶点的重要性。差异分析显示,OGT、GALNT2、GALNT6、GALNT7、PIGT、FUT8、POFUT1、ALG2和UGGT1在ICC组织中的表达水平高于正常组织。生存分析显示,只有FUT8与患者的总生存期(OS)和无复发生存期(RFS)显著相关。SFT治疗不会对FUT8的mRNA或蛋白水平产生显著影响。研究人员通过将这10个残基替换为ALA构建了突变型FUT8,并纯化了FUT8WT和FUT8MT蛋白。MST数据表明,SFT与FUT8WT蛋白具有很强的亲和力,但与FUT8MT蛋白的亲和力降低,Kd值分别为9.83E-08和2.83E-07。模拟对接数据表明,SFT与SER245、ASN247、GLY253、THR267形成稳定的氢键相互作用,与ILE242、TRP255、PRO261形成疏水相互作用。因此,SFT可能直接与ICC-TRC中的FUT8结合。
为了研究FUT8在ICC-TRCs中的作用,研究人员使用特异性的sgRNAs在HUCCT1-TRCs中敲低FUT8。FUT8的缺失显著抑制了HUCCT1-TRCs的克隆球体形成、迁移和侵袭,降低了F-肌动蛋白荧光强度和F/G-肌动蛋白比率,并降低了HUCCT1-TRCs中糖基化的PSGL1蛋白的表达。此外,研究人员将空载体、FUT8WT或FUT8MT质粒转染到FUT8敲除的HUCCT1-TRCs中。研究人员发现,SFT治疗只能将PSGL1从约70 kDa降至约35 kDa,而在FUT8敲除的HUCCT1-TRCs或FUT8MT HUCCT1-TRCs中,效果不明显。此外,SFT治疗在FUT8WT HUCCT1-TRCs中产生了最强的抑制作用,包括克隆球体形成、迁移和侵袭,以及F/G-肌动蛋白比率,而在FUT8KO HUCCT1-TRCs或FUT8MT HUCCT1-TRCs中产生了较弱的抑制作用。综上所述,抑制FUT8的活性是SFT抑制ICC-TRCs的另一种机制。
为了研究FUT8在ICC-TRCs中的作用,研究人员使用特异性的sgRNAs在HUCCT1-TRCs中敲低FUT8。FUT8的缺失显著抑制了HUCCT1-TRCs的克隆球体形成、迁移和侵袭,降低了F-肌动蛋白荧光强度和F/G-肌动蛋白比率,并降低了HUCCT1-TRCs中糖基化的PSGL1蛋白的表达。此外,研究人员将空载体、FUT8WT或FUT8MT质粒转染到FUT8敲除的HUCCT1-TRCs中。研究人员发现,SFT治疗只能将PSGL1从约70 kDa降至约35 kDa,而在FUT8敲除的HUCCT1-TRCs或FUT8MT HUCCT1-TRCs中,效果不明显。此外,SFT治疗在FUT8WT HUCCT1-TRCs中产生了最强的抑制作用,包括克隆球体形成、迁移和侵袭,以及F/G-肌动蛋白比率,而在FUT8KO HUCCT1-TRCs或FUT8MT HUCCT1-TRCs中产生了较弱的抑制作用。综上所述,抑制FUT8的活性是SFT抑制ICC-TRCs的另一种机制。
结语
结语
03
03
本研究揭示了P-选择素/PSGL1轴在ICC中通过重塑细胞骨架促进干细胞特性的新作用。研究人员还发现了硫福罗汀选择性抑制ICC-TRCs的新机制。这些发现扩大了硫福罗汀在癌症治疗中的潜在应用。(转化医学网360zhyx.com)
本研究揭示了P-选择素/PSGL1轴在ICC中通过重塑细胞骨架促进干细胞特性的新作用。研究人员还发现了硫福罗汀选择性抑制ICC-TRCs的新机制。这些发现扩大了硫福罗汀在癌症治疗中的潜在应用。(转化医学网360zhyx.com)
【参考资料】
【参考资料】
https://onlinelibrary-wiley-com.libproxy1.nus.edu.sg/doi/full/10.1002/advs.202407519
https://onlinelibrary-wiley-com.libproxy1.nus.edu.sg/doi/full/10.1002/advs.202407519
【关于投稿】
【关于投稿】
转化医学网(360zhyx.com)是转化医学核心门户,旨在推动基础研究、临床诊疗和产业的发展,核心内容涵盖组学、检验、免疫、肿瘤、心血管、糖尿病等。如您有最新的研究内容发表,欢迎联系我们进行免费报道(公众号菜单栏-在线客服联系),我们的理念:内容创造价值,转化铸就未来!
转化医学网(360zhyx.com)是转化医学核心门户,旨在推动基础研究、临床诊疗和产业的发展,核心内容涵盖组学、检验、免疫、肿瘤、心血管、糖尿病等。如您有最新的研究内容发表,欢迎联系我们进行免费报道(公众号菜单栏-在线客服联系),我们的理念:内容创造价值,转化铸就未来!
转化医学网(360zhyx.com)发布的文章旨在介绍前沿医学研究进展,不能作为治疗方案使用;如需获得健康指导,请至正规医院就诊。
转化医学网(360zhyx.com)发布的文章旨在介绍前沿医学研究进展,不能作为治疗方案使用;如需获得健康指导,请至正规医院就诊。

临床结果临床研究
2024-09-06
欢迎关注凯莱英药闻
(收集周期:9.2-9.6,国内部分为首次申请临床、首次申请上市的创新药)
国内创新药IND汇总
1、和誉生物:ABSK043片
作用机制:PD-L1小分子抑制剂
适应症:肿瘤
9月4日,和誉生物的ABSK043片的临床试验申请(IND)获CDE受理。ABSK043是和誉生物独立自主研发的口服PD-L1小分子抑制剂,是国内首个、全球第二个发布疗效数据的口服PD-L1小分子。与抗体相比,小分子的免疫原性风险低,剂量调节更便捷,患者口服可减少因静脉输注而产生的医疗住院负担。基于小分子的优势和良好的安全性以及初步疗效结果,ABSK043有望成为Best-in-class,为更多肿瘤患者带来临床获益。
根据在2023 ESCO公布的数据:在针对多种晚期实体瘤的临床一期爬坡试验中,ABSK043在11例疗效可评估的BID给药患者中的ORR高达27.3%,且所有剂量组均未发生剂量限制性毒性事件或发现周围神经病变事件。
2、阿斯利康/诚益生物:AZD5004薄膜包衣片
作用机制:GLP-1RA 激动剂
适应症:减肥、降糖
9月4日,阿斯利康/诚益生物开发的AZD5004薄膜包衣片的IND获CDE受理。AZD5004
(ECC5004) 是一种口服GLP-1RA 激动剂,用于治疗肥胖症、2 型糖尿病和其他心脏代谢疾病。该药物既可以作为单药治疗,也可以与针对肠道激素胰淀素的抗肥胖药物AZD6234联合使用。2023年11月,阿斯利康与诚益生物签署了一项独家许可协议,合作开发GLP-1药物ECC5004。在美国开展的1期临床试验显示,与安慰剂相比,在10 mg与30 mg剂量下,药物展现积极的减重与血糖降低效果,且具有>70%的口服生物利用度以及良好的耐受性。
3、阿斯利康:AZD8630吸入剂
作用机制:TSLP单抗
适应症:哮喘
9月4日,阿斯利康的AZD8630吸入剂的IND获CDE受理。AZD8630(AMG
104)是一款抗胸腺基质淋巴细胞生成素(TSLP) 单克隆抗体,有望实现每日一次吸入。TSLP是一种主要由气道上皮表达的细胞因子,在环境损伤时释放,引发一系列下游炎症过程。有充分证据表明,它能够分化2型细胞中幼稚的CD4+T淋巴细胞,产生IL-4、IL-5、IL-13,并降低与1型细胞相关的γ干扰素(IFN-γ)的表达,对多种细胞产生影响。文献表明TSLP信号异常与其他炎症性变态反应性疾病密切相关,包括特应性皮炎、嗜酸性食管炎、慢性阻塞性肺疾病和特发性肺纤维化。此外,它的表达也被病毒、细菌和寄生虫病原体的感染所触发。
I期试验显示,AZD8630每日一次给药,在评估的剂量下长达 28 天内给药是安全的且耐受性良好。接受高剂量 AZD8630治疗的患者 FeNO 显著降低(与安慰剂相比降低 23%, p =0.037);这种降低从 7 天开始明显,持续到治疗期结束(28 天)。大多数不良事件是轻微的,A 部分或 B 部分均未发生严重不良事件。PK 在健康和哮喘受试者中呈线性且一致,支持每日一次给药。
4、科伦博泰:SKB571
作用机制:——
适应症:肿瘤
9月5日,科伦博泰的注射用SKB571的IND获CDE受理。SKB571是一款创新双抗ADC,目前处于临床前的开发阶段,具体靶点尚不明确;主要针对肺癌、消化道肿瘤等多种实体瘤。该药物通过科学的靶点组合选择和差异化的双抗分子设计,提升对肿瘤的靶向性,并有利于克服肿瘤异质性,提升疗效;通过搭配OptiDCTM平台的高亲水性毒素-连接子策略,不仅DAR值均一,且展现了良好的体内药代动力学性质。临床前研究显示,该产品在多种人源肿瘤异种移植(PDX)模型和食蟹猴中,分别展现了良好的抗肿瘤效果及安全性。
2024年8月,科伦博泰宣布与默沙东将就SKB571项目行使独家选择权;默沙东向科伦博泰支付3,750万美元,且待达致特定开发及销售里程碑后,向科伦博泰支付进一步里程碑付款,以及商业化分级特许权使用费等。
5、Galderma:曲法罗汀乳膏
作用机制:RARγ激动剂
适应症:痤疮
9月5日,Galderma的曲法罗汀乳膏的IND获CDE受理。曲法罗汀(Trifarotene)是新一代维A酸类药物,被FDA批准用于治疗面部和躯干痤疮。该药物高选择作用于视黄酸受体γ(RAR-γ) ,RAR-γ是皮肤中主要的RAR亚型;当曲法罗汀选择性发挥疗效的同时,减少了对其他RAR亚型的影响,有效降低了靶器官副作用和皮肤刺激,能以更低的浓度达到同样的治疗效果。大规模 III 期研究PERFECT 1和PERFECT
2 证明,曲法罗汀最早在使用第1周即可有效减少面部病变。Satisfy研究结果显示,从第12周到第52周,曲法罗汀使面部IGA改善由26.6%增加到65.1%,躯干PGA改善从38.6%增加到66.9%。整体成功率57.9%。
国内创新药NDA汇总
1、步长生物:注射用艾帕依泊汀α
作用机制:EPO-Fc融合蛋白
适应症:贫血
9月3日,步长生物的注射用艾帕依泊汀α的上市申请获CDE受理。艾帕依泊汀α是一款促红细胞生成素-Fc(EPO-Fc)融合蛋白,采用的是Fc蛋白融合技术,属于长效 红细胞生成素(EPO) 药物。根据《中国肾性贫血诊治临床实践指南2021》,治疗肾性贫血的药物共有4 种,分为一代、二代和三代EPO,一代为短效EPO,二代和三代均为长效EPO。一代短效EPO 总体的市场规模保持相对稳定,二代长效EPO 2020 年首次国内上市后,市场规模尚未打开,但在2023 年年初被首次纳入医保,预计有望提升市场规模,三代超长效EPO 2018 年国内获批上市,但一直未在国内启动商业化。
公司开展了一项多中心、随机、开放、阳性药对照的 III 期临床研究,旨在施行血液透析的中国慢性肾脏病贫血患者中,比较艾帕依泊汀α和重组 EPO 阿法依泊汀用于维持治疗的有效性和安全性。该试验已经于 2023 年 4 月完成试验,但是试验结果尚未公布。
2、诺和诺德:帕西生长素注射液
作用机制:GH类似物
适应症:生长激素缺乏症
9月5日,诺和诺德的帕西生长素注射液的上市申请获CDE受理。帕西生长素(Somapacitan)为每周一次的长效生长激素,2020年获FDA批准用于治疗成人生长激素缺乏症,商品名为Sogroya;2023年4月将适应症扩大到2.5岁及以上儿童人群。在一项多中心、开放标签、阳性对照、平行组3期REAL4试验中,每周一次接受somapacitan治疗的患者年化生长高度为11.2cm/年。此外,该药物在儿童身高年化生长速度上非劣效于norditropin,未来可与norditropin形成产品组合,满足各类不同患者的需求。目前在美国,Sogroya的10mg/1.5ml规格售价为2496美金(约合1.77万元)。
全球新药III期临床汇总
感谢关注、转发,转载授权、加行业交流群,请加管理员微信号“hxsjjf1618”。
“在看”点一下
临床申请临床2期临床1期申请上市临床结果
2024-07-08
SPRING, Texas, July 08, 2024 (GLOBE NEWSWIRE) --Io Therapeutics Inc., presented today results from studies done in breast, lung, colorectal, and prostate cancer models with the company’s newest anti-cancer compound IRX5010, an agonist of retinoic acid nuclear receptor gamma which was discovered in the company’s oncology drug program. The presentation titled “RAR Gamma Agonist Compounds Promote Effector Memory Tumor Infiltrating T-cells and are Effective in Multiple Cancer Models”, which was delivered at the Federation of American Societies for Experimental Biology (FASEB) Seventh International Conference on Retinoids, being held in St. Paul, Minnesota, USA; was authored by Vidyasagar Vuligonda, Ph.D., Chief Science Officer of the company and inventor of the compound; and Martin E. Sanders, M.D., the company’s Chief Executive Officer.
The presented studies showed that oral treatment with IRX5010 resulted in suppression of growth in mouse models of each of five types of the most prevalent and deadly cancers in humans, i.e. triple negative breast, Her-2 positive breast, non-small cell lung, colorectal, and prostate cancers. In all five models, treatment with IRX5010 induced tumor infiltrating effector memory phenotype T-lymphocytes associated with reduction in tumor growth. Treatment with IRX5010 did not directly impact growth of cancers cells in tissue cultures, indicating that the compound is not significantly directly toxic to cancer cells. The data support that the compound’s mechanism of anti-tumor activity is induction of cancer suppressive tumor-infiltrating T-lymphocytes, so called TILs, rather than direct toxic effects on the cancer cells. Orally administered drugs which induce TILs as a mechanism of action for treatment of cancers are being widely sought in the pharmaceutical industry.
Dr. Vuligonda stated, “These results disclose a new approach to potentially improving treatment of multiple types of serious cancers in humans using IRX5010 as monotherapy, or potentially in combination with other agents such as biologic checkpoint inhibitors, which also induce anti-cancer TIL responses. We are currently conducting preclinical combination studies of IRX5010 with other agents. We anticipate advancing IRX5010 as monotherapy, and as combination therapy other already available treatments, into clinical trials in multiple types of cancers.” Dr. Sanders stated, “IRX5010 has potential to be a new effective oral immunotherapeutic treatment for many types of currently inadequately treated human cancers, including the most common types of mortal cancers. In addition to IRX5010, Io Therapeutics has discovered multiple families of compounds targeting other retinoic acid nuclear receptors, which the company is developing as potential treatments for cancers, neurodegenerative, and autoimmune diseases.”
About Io Therapeutics: Io Therapeutics, Inc. is a privately held company headquartered in Spring, Texas. More information on Io Therapeutics and its product development programs is available on the company’s web site:
Forward Looking Statements: This new release contains "forward-looking statements" within the meaning of the safe harbor provisions of the United States Private Securities Litigation Reform Act of 1995.
Contact:
info@io-therapeutics.com
AACR会议免疫疗法
分析
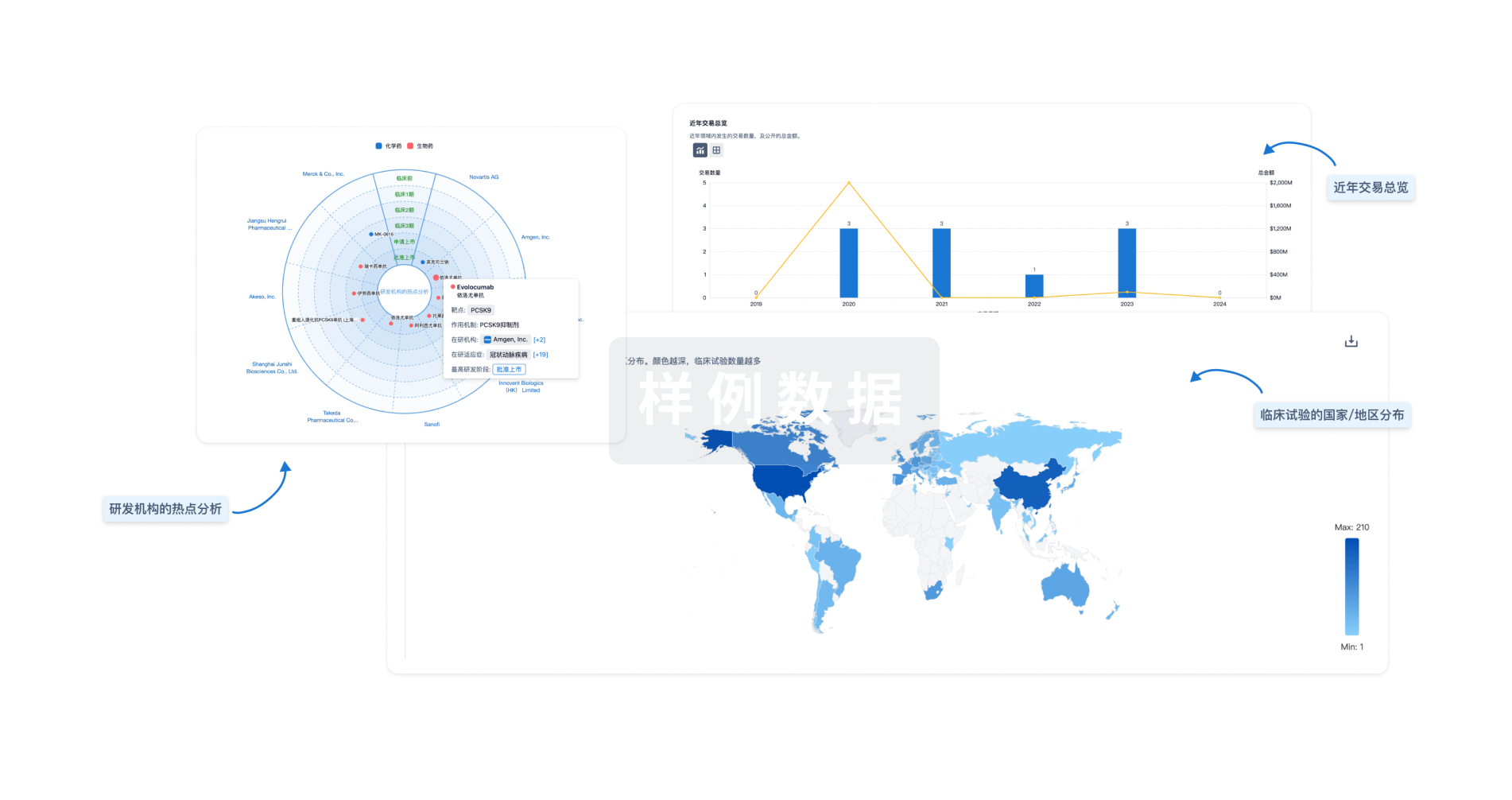
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用




