预约演示
更新于:2025-08-29

Zhejiang University of Technology
更新于:2025-08-29
概览
标签
肿瘤
消化系统疾病
其他疾病
小分子化药
单克隆抗体
融合蛋白
疾病领域得分
一眼洞穿机构专注的疾病领域
暂无数据
技术平台
公司药物应用最多的技术
暂无数据
靶点
公司最常开发的靶点
暂无数据
| 排名前五的药物类型 | 数量 |
|---|---|
| 小分子化药 | 24 |
| 融合蛋白 | 2 |
| 单克隆抗体 | 2 |
| 合成多肽 | 1 |
| 蛋白水解靶向嵌合体(PROTAC) | 1 |
关联
30
项与 浙江工业大学 相关的药物靶点 |
作用机制 ME3 modulators |
在研机构 |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
1
项与 浙江工业大学 相关的临床试验CTR20192025
评价松葛降尿酸颗粒治疗高尿酸血症(湿热蕴结证\痰浊阻滞证)有效性及安全性随机、双盲Ⅱa期临床研究
初步评价松葛降尿酸颗粒治疗高尿酸血症(湿热蕴结证\痰浊阻滞证)的有效性和安全性。
开始日期2019-11-01 |
申办/合作机构  宁波泰康红豆杉生物工程有限公司 宁波泰康红豆杉生物工程有限公司 [+2] |
100 项与 浙江工业大学 相关的临床结果
登录后查看更多信息
0 项与 浙江工业大学 相关的专利(医药)
登录后查看更多信息
19,900
项与 浙江工业大学 相关的文献(医药)2026-01-01·TALANTA
Specific distinguishing between miRNA and pre-miRNA by blocker displacement SMOS-qPCR
Article
作者: Xue, Ying ; Dai, Yanmiao ; Xia, Chenjing ; Xu, Hongwei ; Zhao, Guodong ; Zhou, Xiaojin
MicroRNAs (miRNAs) play crucial roles in various biological processes, and their dysregulation is associated with numerous diseases. Accurate quantification of mature miRNAs is essential for their use as biomarkers. However, distinguishing between mature miRNAs and pre-miRNAs remains challenging. This study introduces a novel method called Blocker Displacement SMOS-qPCR (BL-SMOS-qPCR) to enhance the differentiation between miRNAs and pre-miRNAs. The method employs a blocker sequence complementary to the 3' end of pre-miRNA cDNA, effectively competing with the Linker sequence and reducing non-specific amplification. We optimized various parameters, including blocker modification, length, concentration, and reaction temperature. Results showed that MGB-modified blockers at optimal concentrations significantly improved discrimination between miRNAs and pre-miRNAs, reducing pre-miRNA signals by approximately 3.2-fold. The BL-SMOS-qPCR maintained similar dynamic ranges (6˟108 to 6˟101 copies per reaction) and R2 values compared to the original SMOS-qPCR method across multiple miRNA targets. Furthermore, the new method successfully distinguished between normal controls and esophageal cancer patients in serum samples, demonstrating its effectiveness in clinical applications. This study provides a novel approach for precise miRNA quantification, addressing the challenges of differentiating between mature miRNAs and their precursors in complex biological samples.
2026-01-01·SPECTROCHIMICA ACTA PART A-MOLECULAR AND BIOMOLECULAR SPECTROSCOPY
Aldehydes and ketone sensitive fluorescence sensor constructed by hydroxylamine hydrochloride and carbon quantum dots for baijiu recognition
Article
作者: Chen, Wenxin ; Fan, Yao ; Fu, Haiyan ; Cai, Zhenli ; Wang, Xuhui ; She, Yuanbin ; Fu, Jiahao ; Liu, Yaqi
In this study, a three-channel fluorescence sensor was developed by integrating hydroxylamine hydrochloride (NH₂OH·HCl) with three novel carbon quantum dots (CDs: m-AP@CDs, m-PD@CDs, and o-PD@CDs) to enable the identification of 16 aldehydes, ketones, and diverse baijiu samples. The three-channel sensor was first evaluated for its fluorescence response to aldehydes and ketones, revealing a response intensity hierarchy, ranked as follows: aromatic aldehydes, aliphatic aldehydes, diketones, aromatic ketones, monoketones, and acetal. With the aid of the Linear Discriminant Analysis (LDA) model, all 16 analytes at even low concentrations (2.0 × 10-2 - 2 mmol/L) could achieve 100 % discrimination accuracy. Furthermore, the spliced spectra of the sensor combined with the Partial Least Squares Regression (PLSR) model was used to accurately quantify 16 kinds of aldehydes and ketones in the range of 2.0 × 10-2 - 10 mmol/L. What's more, variations in aldehyde/ketone content across baijiu samples could generate cross-responses with the three-channel sensor, which ensured the sensor could accurately distinguish different flavors, brands and quality baijiu combined with the LDA model, underscoring its potential for rapid, on-site quality control and counterfeit detection in the baijiu industry.
2025-12-01·SPECTROCHIMICA ACTA PART A-MOLECULAR AND BIOMOLECULAR SPECTROSCOPY
Rapid visual authentication of high-temperature Daqu Baijiu using porphyrin signal amplification and smartphone-based cloud machine learning
Article
作者: Su, Yuanyuan ; Long, Wanjun ; Zhu, Yanmei ; Jiang, Xue ; Zeng, Xueqing ; Cang, Yipeng ; Fu, Haiyan ; Lan, Wei ; She, Yuanbin ; Chen, Hengye
High-temperature Daqu Baijiu is known for its superior flavor and value. However, the adulteration of high-grade bottled low-grade Baijiu has become a significant concern. To enhance detection portability, a multi-channel visual sensor array was developed using metalloporphyrin pseudo-peroxidase activity to authenticate the same flavor type Baijiu. The competitive coordination between pyrazine nitrogen atoms and metal ions in porphyrins, along with π-π stacking between aromatic and porphyrin rings, inhibits the pseudo-peroxidase activity of metal porphyrins, leading to 3,3',5,5'-tetramethylbenzidine (TMB) color changes and amplified spectral signals. Combining the colorimetric array sensor with the random forest (RF) algorithm, compared with a single sensor, it can significantly improve the classification accuracy of Baijiu. Among them, sensor points such as TPP_Mg, TPP_Zn, TPP_Cu, and TPP have made outstanding contributions to the identification of Baijiu. In addition, a DD-SIMCA model for identifying adulteration was constructed, and the recognition accuracy rate reached more than 99%. Another, an original smartphone app based on machine learning cloud algorithm was developed for external method verification, and its recognition accuracy rate reached more than 96.83%. Finally, through random forest regression (RFR) analysis combined with the color response of compounds, it was found that Maillard reaction products such as pyrazines, aldehydes and ketones made significant contributions to the identification and classification of Baijiu. The deviation between the actual value and the predicted value of the compound content predicted by this method is less than 0.2473% ± 1.0785%. This study offers a foundation for developing "instrument-free" rapid visual detection methods for high-temperature Daqu Baijiu.
105
项与 浙江工业大学 相关的新闻(医药)2025-08-16
2025年8月15日,“阿维菌素产学研国际联盟” (以下简称联盟)核心成员:中国科学院微生物研究所王为善研究员团队与华东理工大学张立新教授团队、中国农业科学院植物保护研究所李珊珊研究员团队、以及河北兴柏药业合作在线发表了Nature Biotechnology长篇科研论文(article),系统开发出具有优越杀线虫活性的新型阿维菌素类药物柏威霉素(Baiweimectin),并实现了高纯度生物制造。
合作团队进一步开发出专一的、拥有自主知识产权的高纯度生产菌株,其发酵产量达到8.4 g/L(120吨规模),柏威霉素系列衍生物也在持续研发中。
目前,柏威霉素正在国内外同步申报农药登记证。我国每年因线虫病害造成的经济损失高达800亿元,柏威霉素作为拥有自主知识产权的专杀线虫生物农药,对保障农业生产具有重要意义。
01
阿维菌素产业意义与联盟背景
阿维菌素类原料药是由微生物发酵生产的高效低毒生物杀虫剂,对保证我国粮食、农产品安全、畜牧业和医药健康具有重大意义,2亿非洲人也因中国生产的阿维菌素原料药而幸免河盲症。
2024年,阿维菌素全球市场已达13.71亿美元。另外,阿维菌素类新型药物伊维菌素和多拉菌素全球需求增长迅猛,2024年全球市场规模分别高达33.3亿美元和1.34亿美元。
阿维菌素科研攻关的难点在于大规模产业化,中国的高校院所和企业科研人员密切合作,优势互补,最终实现了阿维菌素的高效生物合成,从而大幅度提高了底物利用率、目标产物得率和反应器时空产率,实现了掌控自主知识产权和全球范围产业化,对于提升我国阿维菌素生产企业整体技术水平起到了很好的引领作用。
为进一步聚合行业产学研单位在科研、技术、资金、服务等方面的优势资源,共同促进微生物源生物农药和医药产业的发展,提升产业创新水平和竞争力,带动生物农药的推广应用, 联盟启动仪式于2018年10月12日在青岛隆重举行。
02
联盟成立与发展历程
联盟的成立得到了国内外学术界的关注和祝福,特别收到了曾为阿维菌素产业化做出过杰出贡献的德高望重的三位前辈(浙江工业大学沈寅初院士、中国农业大学李季伦院士以及诺贝尔奖获得者、中国工程院外籍院士、日本科学家大村智教授)的贺信。他们传达了学术前辈们对联盟正式成立热烈而美好的祝福、殷切的期望和深情的鼓励。北京化工大学谭天伟院士和华东理工大学张嗣良教授等专家、学者、企业领导上台共同按下启动球,一起见证了联盟的启动。
联盟集结了生物反应器工程、微生物代谢、微生物资源前期开发、分子反应动力学、农业微生物学共5个国家重点实验室,1个国家生化工程研究中心和1个省部级微生物活性产物工程研究中心,还有阿维菌素发酵行业内的多家龙头企业,针对阿维菌素耐药性机制、衍生物生成原理以及智能化高效生产的策略等行业巨大需求展开联合攻关。美国默克公司于上世纪80年代率先将阿维菌素推向市场。
为满足我国农业需求,沈寅初院士和李季伦院士同步在国内展开相关研究,并分别在1999年和2006年两次获得国家科技进步二等奖,支撑了四家龙头企业的产业化和成功上市。联盟共同主席张立新教授组建攻关团队,利用合成生物学技术实现阿维菌素生物制造的最高产量(9.3 g/L),该技术转移到三家龙头企业(威远生化、河北兴柏药业、齐鲁制药),2016年联合获得国家科技进步二等奖,2020年在Nature Biotechnology发表长篇科研论文(doi:10.1038/s41587-019-0335-4),同时申请了包括PCT在内的多项专利,近三年新增销售额达25.8亿元,新增利润6.22亿元。
03
最新成果与技术突破
在这项研究中,张立新教授组织团队(包括中科院微生物所王为善研究员、中国农业科学院植物保护研究所李珊珊研究员、华东理工大学张敬宇副教授等)与企业合作,开发了一套普适性重编程工程策略(下图):
链霉菌高产菌种重编程技术体系
1.构建智能控制系统: 基于对链霉菌群体感应控制系统多样性及信号传导过程的深入理解,开发了普适于该物种的正交、多路人工动态控制系统——SMARTS智能体;
2.提出适配原则与流程: 利用链霉菌次级代谢共性特征,提出了一套多靶点适配原则与标准化工作流程;
3.创制高产菌株实现产业化: 应用上述原则,成功开发出用于生产新型抗线虫药物柏威霉素的阿维链霉菌高产菌株,并在120立方米的工业发酵规模上实现了高效生产。
该技术体系为链霉菌次级代谢药物的高效生物制造提供了核心支撑。值得一提的是,近年来,中国科学家曾在Nature Biotechnology期刊上发表了三篇微生物合成领域的研究长文,都来自同一个团队,这也是该团队获2016年度国家科技进步二等奖“阿维菌素的微生物高效合成及其生物制造”项目的进一步提升。
04
未来展望
基于以上积累,联盟目前正在聚焦新型阿维菌素类药物伊维菌素(Ivermectin)、多拉菌素(Doramectin)、依立菌素(Eprinomectin)、米尔贝霉素(Milbemycin)、莫西菌素(Moxidectin)等进行生物制造技术创新,力争抢占该类药物生物制造的制高点。
阿维菌素类原料药智能生物制造的成功范例也将为我国其它活性天然产物智造、品种改良和节能减排、产业化升级提供有益的理论、思路和方法。
赵国屏院士点评
合成生物学是以工程化的思维认识生命体系,为我们对生命本质的理解提供了新的视角。合成生物学研究的一个重要方向,是“将生命过程作工程化设计重构”—“建物致用”赋能生物工程—特别是代谢工程,实现对复杂系统有效且可复制的定量控制,被认为是引领下一次产业革命的关键技术之一。
工业微生物由于“底盘”类型和代谢产物的多样性以及调控网络的复杂性,常被视为难以实现“通用化”改造。因此通过系统生物学研究,以合成生物学思路理性设计超越自然的人工调控元件/模块,是实现工程化改造工业微生物的重要途径。张立新、王为善和李珊珊研究团队基于对链霉菌特有群体感应系统的进化分析,发现虽然链霉菌群体感应信号多样,然而趋异进化的受体蛋白却趋同识别同一DNA结合位点。利用这一新认识开发普遍适用于该物种的动态多路人工控制系统(SMARTS),可在多种链霉菌中动态响应不同的群体感应信号,并辅以多靶标适配策略实现了新型抗线虫药物柏威霉素和抗肿瘤药物表柔比星的高效生产,且在产业化应用中保持鲁棒性。该工作系统认识链霉菌群体感应,总结进化的个性与共性,人工构建一套使二者协同的使能工具,实现在不同菌种中的“通用性”应用。
尽管工业微生物“黑箱”难以被彻底认识,但生物进化的规律往往存在可预测性,可被研究人员进一步挖掘,进化为合成生物学提供“设计蓝图”,而合成生物学为进化提供“验证平台”,该工作通过对进化这一底层规律的认知,形成了次级代谢多目标协同重编程的原创技术体系,体现了进化认识与合成生物学研究理念相结合的应用潜力,为推动工业微生物从“致知”到“致用”的研究提供了借鉴。
邓子新院士点评
细菌群体感应系统作为一种单细胞生物个体之间发生协同性群体行为的调控模式,是近年来微生物学领域的重大发现之一。链霉菌属群体感应系统普遍性的调控了次级代谢的开启,然而对该物种群体感应的认识仅是冰山一角。因此,研究链霉菌群体感应系统不仅有助于理解次级代谢产生规律,还将指导动态控制系统重构及其高产菌株开发。张立新、王为善和李珊珊研究团队发现链霉菌群体感应受体蛋白与结合启动子进化的趋同性,基于这一新认识开发普遍适用于该物种的动态多路人工控制系统(SMARTS)。通过巧妙的合成生物学电路设计,克服链霉菌群体感应信号瞬时性开启的难题,且正交的控制与次级代谢高产的多个靶点而不与链霉菌本源网络串扰。利用SMARTS与多靶点适配原则实现了新型抗线虫药物柏威霉素和抗肿瘤药物表柔比星的高效生产,特别是推动了柏威霉素的产业化应用。该工作从底层基础认识到创新技术再到产业化应用,开发人工链霉菌群体感应系统,服务于链霉菌高产菌株开发,是我国链霉菌来源的药物高效生物制造的成功实践。
该工作是继经典革兰氏阴性菌和阳性菌的群体感应研究后,对链霉菌这一重要微生物来源药物物种群体感应的系统认识,并基于全新认识,开发的人工多路动态控制系统,实现在不同链霉菌的即插即用和正交控制,是赋能链霉菌次级代谢药物高效生物制造理想工具。该工作同时也为其它天然信号系统的工程化开发提供借鉴。
钱旭红院士的点评
生物制造推动了全球新一轮科技革命与产业变革,其中微生物来源药物的生物制造已成为各国科技竞争的战略制高点之一。微生物天然产物农药因其生物活性强、靶向性高及环境兼容性优良,作为绿色农药的重要组成部分,在保障国家粮食安全和农产品质量安全方面具有重要意义。链霉菌作为天然产物农药的关键生产菌株,长期面临发酵效价低、靶点改造效率不足及菌株优化试错率高等挑战。因此,建立一种普适性的链霉菌多靶点协同高产改造策略显得尤为重要。
针对上述问题,张立新、王为善、李珊珊等基于对链霉菌代谢调控规律的系统认知,开发了一种适用于该物种的动态多路人工控制系统(SMARTS),并据此构建了链霉菌多靶点协同优化的高效菌株改造策略。该策略成功实现了新型抗线虫药物柏威霉素及抗肿瘤药物表柔比星的高产菌株构建,其中柏威霉素已实现规模化生产(规模达120吨,产量8.4 g/L)与大田实验应用。
绿色农药的大规模推广依赖于高效的量产能力。如何实现农用天然产物的高效生物制造,成为亟待解决的问题。该研究为这一问题提供了可借鉴的解决方案。具体而言,研究团队通过人工控制系统对菌株进行智能重编程,实现了菌株的智能可控性,并与生产工艺高度匹配。由此,开创了智能控制人工重编程技术与生物制造过程相结合的新路径,为链霉菌天然产物农药的高效生物制造及创新应用提供了切实可行的“智能制造”方案。
张克勤院士的点评
合成生物学通过将生物系统转化为可编程的细胞工厂,将代谢工程从试错式改造升级为理性设计与智能优化的精密科学,多靶点适配是合成生物学从改造“单个零件”迈向构建“复杂机器””乃至“智能系统”的必由之路。张立新、王为善和李珊珊研究团队开发普遍适用于链霉菌属的动态多路人工控制系统(SMARTS),进一步基于次级代谢的共性规律,开发多靶点适配原则和标准化工作流程,首先多目标优化直接参与合成机器构成及其分流的靶标,实现多酶高效协同生产;然后进一步适配支撑次级代谢合成的相关靶点,实现新型抗线虫药物柏威霉素和抗肿瘤药物表柔比星的重编程生产。值得一提的是,在工业级别的放大生产中,SMARTS与多靶点适配原则仍能动态同步开启并协同控制多个与高产相关的靶点,精准控制代谢通量,提升了工业链霉菌的鲁棒性与稳定性。
微生物来源药物的高产往往需要多轮迭代,且易陷入局部最优的困境,传统代谢工程往往采用经验驱动的“高、中、低”组合试错,该工作引入工程学“多目标优化”的适配策略,实现多个靶点更精细的组合,从而使其协同一致的为次级代谢生产服务。该工作为其它复杂天然产物药物的适配工程提供理论指导与技术支撑,也将推动定量合成生物学指导下高产菌种开发的发展。
原文链接:https://www-nature-com.libproxy1.nus.edu.sg/articles/s41587-025-02762-1
识别微信二维码,添加生物制品圈小编,符合条件者即可加入
生物制品微信群!
请注明:姓名+研究方向!
版
权
声
明
本公众号所有转载文章系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系(cbplib@163.com),我们将立即进行删除处理。所有文章仅代表作者观不本站。
微生物疗法
2025-08-12
·中国药闻
近日,各地药监部门采取多种形式,主动靠前服务,助力医药产业高质量发展。
组织座谈交流
为进一步优化医疗器械出口销售证明办理服务,北京市药监局组织召开医疗器械出口企业座谈会,聚焦企业“走出去”需求,研讨政策优化路径。市药监局积极回应企业关切,明确将从强化政策对接、优化服务机制、赋能企业“出海”等方面提升服务效能,以优化医疗器械出口销售证明服务机制为切入点,全力支持企业抢抓全球市场机遇,更好抢占国际市场。
河北省药品职业化检查员总队(南片区)近日组织召开药品生产企业专题帮扶座谈会,技术专家组与企业技术团队及设计公司展开深入探讨,全面梳理企业在技术层面存在的瓶颈问题,给出建设性意见,同时还就具体问题提出改进建议。会议紧扣企业急难愁盼问题,通过“面对面”倾听诉求、“点对点”纾困解难的务实举措,为省内药品生产企业提供权威指导与政策指引,用实际行动践行“监管为民、服务惠企”的宗旨。
浙江省药监局先后赴浙江工业大学、浙江大学开展化妆品原料创新组团服务并组织座谈交流。在浙江工业大学实地参观了化妆品植物原料研究团队实验室,详细了解该团队在特色植物资源开发、活性成分提取与功效评价等方面的研究进展。在浙江大学实地参观了智能医药实验室,听取关于利用人工智能、合成生物学等前沿技术开展化妆品原料创新研发的情况介绍,同时对各研发团队在化妆品原料创新中面临的问题和困难进行了交流和解答。
开展服务日活动
安徽省药监局党组书记、局长马旭升主持开展第十二期“局长服务日”活动,活动现场,省药监局与会人员逐个听取企业意见、建议,对企业提出的药品生产企业兼并重组、药品生产工艺变更、产品持有人变更、抗肿瘤药分段生产等具体问题一一予以回应;对于需要跟进服务的事项,省药监局将做好后续服务与跟踪督办工作。省药监局将坚持寓服务于监管,全面强化药品全生命周期监管,进一步加大政策宣贯与培训力度,助力全省医药产业高质量发展。
江苏省药监局常州检查分局、审评核查常州分中心以企业需求为导向,深化服务举措、拓宽服务覆盖面,持续开展“局长/主任接待日”活动,助力企业突破发展瓶颈,为常州生物医药产业整体高质量发展持续赋能。今年以来“局长/主任接待日”活动已成功举办十余次,覆盖企业数十家。
云南省药监局党组成员、副局长张路晗带队赴楚雄开展2025年第五期局领导政务服务接访日活动,面对面倾听辖区内17家药品生产企业及医疗机构的诉求,精准破解发展难题。此次活动聚焦企业高度关切的《药品生产许可证》《医疗机构制剂许可证》重新发证流程、GMP规范执行等核心问题,省局参会人员逐一进行现场回应与指导;对需跨部门协调的复杂事项,现场记录并承诺及时反馈解决方案,确保企业诉求“件件有回音”。通过“政企直连”模式,切实打通政务服务“最后一公里”。
组织开放日活动
宁夏回族自治区药监局近日举办了“政府开放日”活动,受邀代表参观了药监局业务办理窗口,观摩并体验了执业药师办理过程,现场设置了答疑解惑环节,代表还就如何进一步优化药品监管服务、提升监管效能等提出了宝贵的意见建议。此次“政府开放日”活动是自治区药监局“我陪群众走流程”的重要活动之一,通过邀请群众和企业代表参与多维度、沉浸式体验,强化现场监督效果,助推监管部门持续优化审批流程,让服务更贴心、更高效。
山西省药品审评中心持续深化“药品化妆品咨询开放日”活动,设立“咨询开放日”是省药品审评中心的创新服务模式,通过面对面了解企业诉求,一对一提供精细化、专业化指导服务,实打实帮助企业解决实际难题。活动坚持高频次、常态化举办,今年以来线上线下活动共接待省内药品生产企业40余家次、化妆品企业30余家次,累计解决药品问题60余条、化妆品问题100余条,当场问题解决率超85%。
贵州省药监局检查中心开展“化妆品企业开放日活动”暨推进化妆品安全评估工作会,通报化妆品安全评估工作推进情况,针对企业困难提供技术指导。活动详细解读了《化妆品生产质量管理规范检查要点及判定原则》重点要求,结合前期化妆品企业检查中发现的共性及常见问题进行分析讲解,强化企业对法规规范的认识和理解。最后,针对收集到的30余个问题进行现场答疑,帮助企业解决备案和生产质量管理技术问题。
举办技术培训
上海市器审中心以“优化营商环境”为主线,聚焦企业急难愁盼问题,联合普陀区市场监管局组织开展化妆品备案合规“体检”。培训会上,器审中心介绍了《化妆品备案合规“体检”指引》,重点讲解了《错题自查自纠表》的下载和使用方法,以及如何对照开展自查自纠,建立起符合自身实际的自主合规“体检”机制,形成"自查-整改-提升"的闭环管理,把问题尽可能地解决在产品上市之前。培训最后现场解答了企业关于备案信息修改流程、原料报送码使用、物料委托放行等12项疑难问题。
陕西省商洛市药检所组织省级药品GMP检查员、检验检测专业人员深入企业开展帮扶指导,针对企业提出的中药饮片标签管理、检验报告书写、2025版药典实施等5大类11个方面问题逐一进行解答指导,并就企业在原辅料管理、生产过程质量控制、检验检测等方面存在的问题进行现场教学培训,及时解决了企业在药品生产和质量管理方面存在的问题,进一步提高企业质量管理水平。今年以来,商洛市药检所已开展入企帮扶指导3次,培训药品从业人员65人次,为企业纾困解难13项。
(根据相关省份药监局信息整理)
高管变更一致性评价
2025-08-06
以下文章来源于安徽省科创投资有限公司/安徽国控投资有限公司公众号科创汇投,作者张达
近年来,相关政策对于合成生物产业的支持力度不断加大,各省均在加快布局合成生物产业集群,行业龙头企业纷纷入场,科技型初创企业不断涌现。安徽发展合成生物产业有基础、有亮点,成功培育了华恒生物、丰原集团、安科生物、中盛溯源等行业头部企业,涌现出阿法纳生物、和晨生物、瑞邦生物、通用生物等一批科技型中小企业,丙氨酸、聚乳酸、烟酰胺等产品市场份额和产能处于国内外领先地位,产业发展迸发蓬勃活力,为安徽抢占未来产业创新制高点奠定基础。本篇作为系列研究的中观篇,着重介绍安徽省合成生物产业布局和代表企业概况。
一、安徽省合成生物产业概况
当前国家大力推动“新质生产力”发展,以合成生物技术赋能生物制造,通过技术创新、政策支持与产业投资等举措加快推进生物制造高效可持续发展。《“十四五”生物经济发展规划》明确提出,推动合成生物等前沿技术的产业化,构建具有国际竞争力的生物制造产业体系。安徽省积极响应国家战略,合肥生物制造省未来产业先导区已被列入安徽省十大未来产业先导区之一。
根据安徽省工信厅公开数据,2024年安徽生物制造产业规模约600亿元,占全国比重约5%,逐步形成以合肥、蚌埠为“双核”,皖北、皖江为“两翼”的生物制造产业空间布局。当前安徽合成生物产业主要优势布局在合肥、蚌埠、芜湖、滁州等地市,合成生物关联企业覆盖全省16个地市,以基础研发和初创型科技中小企业为主,合成生物产业作为战略性新兴产业受到相关政府部门高度重视。
图1 安徽合成生物产业重点地市布局
安徽省积极推动合成生物产业发展,加快科技创新与产业转型升级,在企业培育、产业布局、投资赋能、场景应用等方面采取积极措施。相关国有投资机构、政府引导基金联合社会资本加快布局合成生物赛道,通过投资培育本土企业和招引优质企业并举,支持合成生物相关企业在皖落地和做强做大。为积极布局培育生物制造未来产业,安徽省中小企业(专精特新)发展基金充分发挥政府投资基金的引导作用,联合国家开发投资集团、合肥市、芜湖市等共同发起设立规模20亿元的国投安徽生物制造基金,该基金是长三角内首支以合成生物成果转化为特色,聚焦生物制造产业的主题基金。重点投向生物制造产业及生物技术方向,促进合成生物成果转化。
2024年10月,安徽省工业和信息化厅发布《安徽省生物制造产业高质量发展行动方案(2024-2027年)》(征求意见稿),提出到2027年,形成一批具有重要影响力的生物制造科技创新成果,布局一批具有明显经济效益的生物制造产业化项目,引进和培育一批高成长企业,形成较为完备的生物制造赋能医药、化工、材料、食品、能源等产业高质量发展的产业链体系,构建创新活跃、转化顺畅、市场繁荣、要素完备的产业生态格局,带动生物制造相关产业规模突破1000亿元。
表1 近年安徽和地市合成生物相关重要政策梳理
二、安徽省重点地市合成生物产业布局
在政策支持和技术进步的双重驱动下,知名投资机构、创新研发企业纷纷进入合成生物产业赛道,助力产业加速发展。安徽省在生物基材料、生物基化学品、生物医药等细分领域涌现了丰原生物、恒鑫生活、华恒生物、和晨生物、利夫生物、瑞邦生物、中科健康、通用生物、中盛溯源、阿法纳生物、安科生物等包括上市公司在内的合成生物产业链相关企业,同时凯赛生物、华熙生物、微构工场、智飞生物等省外代表性企业也相继在安徽落地。本文对合肥、蚌埠、芜湖、滁州等合成生物产业布局重点地市进行研究。
1.合肥
合肥市将生物制造作为五大先导产业之一,出台生物制造产业高质量发展行动方案,组建合成生物创新研究院,围绕生物制造产业加速布局,形成了集群式发展态势,生物制造产业呈“两核多点”的产业空间布局。合肥市以长丰县和高新区为两核,经开区、蜀山区、巢湖市、肥西县、庐江县多点发展。通过产业规划、项目谋划、资金筹划“三划一体”谋划布局,依托合肥市发改委成立生物制造产业专班,并借助科大硅谷等创新载体,持续深化“双招双引”。同时推进与央企、龙头企业深入合作,三大央企招商局集团、保利集团、国投集团均在合肥围绕合成生物产业布局多元合作或战略投资。2024年合肥市共招引生物制造重点项目10余个、协议投资额近140亿。当前合肥市集聚包括华恒生物、凯赛生物、恒鑫生活、安科生物等龙头企业在内的超40家产业链相关企业并呈快速增长趋势,覆盖先进材料、消费品、生物医药、农业技术等多个领域。
2025年2月27日,合肥生物制造省未来产业先导区、合肥(长丰)合成生物制造产业园正式揭牌,将围绕生物基材料、农业生物育种、生物医药等合成生物产业重点领域布局。长丰县目前已汇聚安徽省合成生物制造产业创新研究院和华恒生物、恒鑫生活、微构工场、昌进生物、康诺生物等重点研发机构和企业,初步形成合成生物产业集群。长丰县率先建立安徽省首个生物制造产业园,确立“南研北制”布局,建设小试、中试、量产、检验检测平台等基地,不断完善上下游产业链,打造省内合成生物产业发展新高地。
合肥市高新区依托生物医药产业发展优势,汇聚凯赛生物、和晨生物、利夫生物、中科健康、安科生物、智飞龙科马、兆科药业、中盛溯源、中科普瑞昇等涵盖生物基材料、生物基化学品、生物医药等合成生物相关企业。2024年8月,高新区生物制造产业园(凯赛生物产业园)已完成备案。
根据2024年5月合肥市发布的《合肥市推进生物制造产业高质量发展行动方案(2024—2026)》,合肥明确提出合成生物制造产业的定位、发展空间、功能布局、支撑体系、实施计划等,未来将突出企业为主体和产学研联动、技术创新和产业应用协同,以示范应用带动技术迭代,丰富拓展新场景新赛道。方案提出力争到2026年,基本建成全链条创新平台体系,打造国内一流生物制造产业园区,培育引进一批具有国际影响力的生物制造领域领先企业。力争到2026年,建设3个以上国内领先的专业产业园区,生物制造产业集群产值规模突破300亿元,形成初具规模业态完整的生物制造产业集群。
表2 合肥市合成生物产业相关企业
2.蚌埠
蚌埠市凭借传统生物发酵和新材料产业基础向合成生物转型,打造以生物基材料为主的生物制造产业集群。蚌埠市大力发展生物基新材料产业,积极推广生物基可降解新材料制品,在应用端通过延伸拓展产业上下游链条,推进生物可降解材料在包装制品、纺织服装、家具用品、生物医药、精细化工等领域的场景应用。作为全国唯一以生物基材料为特色的国家新型工业化产业示范基地,蚌埠汇聚了丰原生物、中粮科技、雪郎生物等一批合成生物产业相关龙头企业。在生物基材料领域,形成了以聚乳酸、聚丁二酸丁二醇酯、呋喃聚酯、聚氨基甲酸酯、纳米纤维素为特色的“四聚一素”产业发展体系,建成发酵技术国家工程研究中心等国家级创新平台7个,集聚生物基新材料产业链企业超70家,产业规模突破200亿,加快培育世界级生物基新材料产业之都。
蚌埠市锚定生物基新材料产业高质量发展目标,以安徽丰原集团等龙头企业为依托,围绕产业链部署创新链,推动生物制造产业能级跃升。丰原集团已在固镇经济开发区建立面积约5300亩的丰原生物产业基地,拥有16家规上企业,投资170多亿元建设生物发酵、生物材料、生物能源及生物肥料等30多个项目。2020年3月,国家工信部公布了第九批国家新型工业化产业示范基地名单,固镇县申报的(生物基新材料)新型工业化产业示范基地名列其中,这是全国首个以生物基新材料为特色的产业示范基地。根据固镇县发改委公开数据报道,2023年固镇县生物基新材料产业实现产值61.04亿元,集聚上下游高新技术企业44家。固镇县聚焦生物基新材料产业,着力打造全球产业规模最大、产业链条最长、产品种类最多的千亿级生物基新材料产业集群。
表3 蚌埠市合成生物产业相关企业
3.芜湖
芜湖市依托生物医药产业基础,坚持走“产业+科创”之路,向合成生物产业延伸发展。芜湖市以三山经济开发区为合成生物产业核心集聚区,同时在鸠江、南陵等县区多点布局、协同发展,汇集了普立思、华仁科技、英特菲尔、瑞驰兰德、万泽等代表企业。根据芜湖市人民政府官方网站数据,芜湖市在合成生物产业及上下游全产业链领域拥有规上企业10家,高新技术企业超20家,全产业链总产值超40亿元。
2024年9月30日芜湖市出台《芜湖市合成生物产业发展行动方案(2024-2027年)》,芜湖市将围绕生物基材料、生物医药、生物农业、生物食品、生物装备制造、生物平台工具等六大发展方向,谋划十项工作举措,包括布局一批重点项目,培育一批种子企业,规划合成生物专业园区等。同时,未来继续围绕“产业+科创”深度融合,加强产业科技创新引导,建设研发中试和创新载体平台,实施关键技术专项攻关,搭建合成生物应用场景,并强化科技金融支持,培养合成生物人才梯队,组建芜湖市合成生物产业联盟,推动创新链、产业链、资金链、人才链深度融合。力争到2027年,转化一批具有产业价值的合成生物科研成果,招引培育拥有产业关键核心技术的合成生物科技型企业100家以上,打造国内领先的合成生物特色产业创新高地。
表4 芜湖市合成生物产业相关企业
4.滁州
滁州市将生命健康产业作为重点打造八大产业链之一,依托滁州市卫健委成立生命健康产业推进专项工作组。通过培育和招引生命健康相关企业50多家,集聚了一批包括“潜在独角兽企业”通用生物、国内最大的β-丙氨酸生产企业华睿生物在内的合成生物相关企业。本土上市公司金禾实业积极布局合成生物赛道,合成生物产品涉及甜菊糖苷、罗汉果甜苷、阿洛酮糖、红没药醇、圆柚酮等食品添加剂和日化香料品类。
滁州市中新苏滁高新区将生命健康产业作为园区两大支柱产业的首要产业,发挥园区现有的生命健康产业基础优势,抢抓合成生物产业发展风口期,致力于打造滁州市合成生物产业核心优势区,加快推进合成生物产业集聚发展。省外合成生物相关企业华睿生物、趣酶生物、斯拜科生物等相继落地中新苏滁高新区。
表5 滁州市合成生物产业相关企业
5.其他
安徽省各地市积极布局合成生物相关产业。其中宿州市高度重视合成生物产业发展,正加快推动合成生物技术在农业、绿色食品、材料等领域的产业化应用,并推动建设合成生物产业园,已集聚新宇药业、新熙盟生物、中农秸美等生物医药、生物能源相关企业。阜阳市拥有博科力生物、福方高科等涵盖食品营养、农业技术等合成生物细分领域相关企业。阜阳市在“6849”产业发展布局构建中,合成生物被纳入前瞻布局的四大未来产业之一。其中颖东区工业园扩区设立生物产业园区,目前已落地多个合成生物科创研发与生产项目。安庆市当前依托双生谷产业园(安庆高新技术产业开发区“双生谷”片区),聚焦生命健康和生物科技两大产业,加速招引合成生物相关企业,多个重点项目如神州种谷、朗坤药业、普利制药、华大基因等正在同步推进。同时双生谷与含元资本、达安基因、复星集团等知名企业合作建立基金,为相关产业落地提供有力支持。马鞍山市着力推进生命健康产业发展,成立生命健康等11个新兴产业推进组,初步形成以瑞邦生物、皓元药业、泰恩康制药等为产业链龙头的合成生物产业布局。慈湖高新区依托皓元医药马鞍山生物医药公共服务平台优质的技术服务及辐射带动功能,重点发展生物医药等产业。当涂经开区全力打造生命健康产业集群,园区内汇聚了瑞邦生物、泰恩康制药等合成生物代表企业,作为国家级专精特新“小巨人”企业,瑞邦生物烟酰胺产品国内市场的占有率稳居行业首位,有望获评国家级制造业单项冠军企业。
三、安徽省合成生物重点企业
1.华恒生物
安徽华恒生物科技股份有限公司(688639.SH)位于安徽合肥,是一家以合成生物技术为核心驱动力的高新技术企业,专注于绿色科技创新和绿色价值创造。公司主要产品涵盖氨基酸、维生素和生物基材料单体等,广泛应用于中间体、动物营养、美妆与护理、植物营养以及功能食品等领域。华恒生物与中科院天津工业生物技术研究所联合研发的发酵法生产L-丙氨酸技术,成功实现了微生物厌氧发酵规模化生产L-丙氨酸。借助成本和绿色低碳优势,华恒生物L-丙氨酸全球市占率高达70%,L-丙氨酸产品在2019年被工信部认定为“制造业单项冠军”产品。华恒生物联合浙江工业大学在长丰县建设高能级合成生物创新研究院,人工智能驱动生物制造研发及中试示范基地建设项目和人工智能精准发酵及蛋白质工程共享示范项目相继落地合肥市长丰县生物制造产业园。
2.凯赛生物
上海凯赛生物技术股份有限公司(688065.SH)是一家以合成生物技术为基础,从事新型生物基材料的研发、生产及销售的高新技术企业。公司目前实现商业化生产的产品主要聚焦聚酰胺产业链,涉及生物基聚酰胺以及可用于生物基聚酰胺生产的原料,包括DC12(月桂二酸)、DC13(巴西酸)等生物法长链二元酸系列产品和生物基戊二胺,是利用生物制造规模化生产新型生物基材料的标杆企业。凯赛生物先后在合肥成立安徽凯酰时代复合材料有限责任公司和招商凯赛复合材料(合肥)有限公司。其中凯酰时代由凯赛生物、宁德时代旗下时代泽远基金以及杭州卡涞复合材料联合成立,凯酰时代依托凯赛生物深厚的技术储备和强大的产业化能力以及宁德时代在动力电池行业的应用场景和市场资源,扩展生物基符合材料在新能源汽车和储能市场的应用。招商凯赛由凯赛生物、招商创科和合肥市政府共同设立,旨在联合布局生物基复合材料建设基地,通过“合肥模式”,构建出具有全球竞争力的合成生物材料产业集群,并建立合成生物学研究和产品应用开发的重要平台。
3.丰原生物
安徽丰原生物技术股份有限公司基于菌种、发酵、提取、纯化、聚合、应用开发六大核心合成生物制造技术,以玉米、木薯、农作物秸秆等为原料,生产主要涵盖三大有机酸和八大氨基酸系列生化产品。丰原生物依托丰原集团强大的创新能力,经过多年的持续攻关,将废弃秸秆由生态包袱发展为生态产业,实现秸秆原料到混合糖到聚乳酸至下游应用到联产黄腐酸有机肥等产业链集群发展,延伸出塑料加工、纺织、工业及家用装饰材料、生物化学品、生物燃料、生物肥料等六大产业集群。丰原生物入选长城战略咨询2024年“独角兽企业”榜单。
4.恒鑫生活
合肥恒鑫生活科技股份有限公司(301501.SZ)成立于安徽长丰双凤经济开发区,是一家致力于环保产品研发、生产和销售的高新技术企业。恒鑫生活是中国聚乳酸(PLA)制品规模化生产的应用典范,在全球PLA快消产品市场占有重要地位。公司践行绿色发展使命,加快生物降解技术产品迭代,推动新材料的多元化应用,以科技创新促进行业可持续发展,公司入选工信部第四批“专精特新小巨人”企业名单。
恒鑫生活与微构工场合作,双方基于聚羟基脂肪酸酯(PHA)与PLA材料的共混改性共同拓展生物可降解材料应用,恒鑫生活已成功荣获中国首张PHA淋膜纸制产品的DIN证书,将有力推进PHA在消费级包装材料的应用场景拓展。
5.微构工场
北京微构工场生物技术有限公司是一家专注于合成生物学与生物制造的高新技术企业,依托自主研发的嗜盐菌底盘体系,开发可自然降解的环保材料PHA,致力于推动绿色生物基材料的产业化应用。微构工场是清华大学科技成果转化企业,先后获评“瞪羚企业”、“潜在独角兽企业”、“专精特新中小企业”等资质。公司先后与安琪酵母(600298.SH)和川宁生物(301301.SZ)成立合资公司建设PHA生产基地。
微构工场与安琪酵母合作在湖北恩施的全国首条年产1万吨合成生物PHA生产线已于近期正式交付,即将正式投产。微构工场在合肥市长丰县成立安徽微构工场合成生物有限公司,致力于生物基材料的研发与生产,主要产品包括可降解材料PHA(聚羟基脂肪酸酯)及相关技术解决方案。安徽微构工场位于长丰双凤经济开发区的绿色材料PHA创新研发与高端智造产业生态园项目正在建设中,预计2027年7月投产。
6.安科生物
安徽安科生物工程(集团)股份有限公司(300009.SZ)位于合肥市高新区,是一家长期致力于生物医药研发、生产、销售和投资的高新技术企业。现有产品与服务涵盖生物制品、现代中药、化药及小分子药物、多肽药物、体外诊断试剂、法医检测、靶向抗肿瘤药物、细胞免疫治疗等众多领域。安科生物重组人生长激素国内市场占有率在15%左右,注射用曲妥珠单抗作为安徽省第一个获批上市的生物大分子抗肿瘤药物,填补了安徽省抗体药物领域的空白。安科生物积极布局创新疗法,控股或参股瀚科迈博、博生吉安科、阿法纳生物、元宋生物等细胞与基因治疗相关创新研发企业,作为基石投资者参股维昇药业(2561.HK)。
7.中盛溯源
安徽中盛溯源生物科技有限公司位于合肥市高新区,由全球著名科学家、人诱导多能干细胞(hiPSC)发明人之一俞君英博士领衔创建。公司专注hiPSC技术研发与临床转化和干细胞药物研发,通过GMP级生产,扎实、稳步推进细胞药物的临床申报工作,为肿瘤治疗、自身免疫性疾病、神经退行性病变等疾病提供创新治疗方案,持续探索hiPSC来源细胞药物的临床价值。中盛溯源系统性建立了围绕hiPSC技术应用的发明专利组合,覆盖了hiPSC的重编程、多种功能细胞的定向诱导分化、生产工艺和关键原材料等上下游。公司推进多个基于hiPSC来源细胞药物进入临床试验阶段,其中NCR100和NCR300为国内首款获批临床的iMSC和iNK细胞产品,NCR101为全球首款获批临床的基因修饰iMSC细胞产品。
8.和晨生物
合肥和晨生物科技有限公司位于合肥市高新区,是一家以合成生物技术驱动的高新技术企业。和晨生物通过高效微生物细胞工厂的构建与优化,实现功能营养成分、生物活性成分等关键功能活性原料的规模生产与绿色制造,业务涵盖菌种工程、发酵、提取工艺、放大量产、产品配方解决方案和市场渠道等领域。公司主要以稀有氨基酸及其衍生物为核心,包括生物基化学品、小品种氨基酸、高附加值活性原料三大原料产品板块。公司正着力打造麦角硫因、茶氨酸、根皮素等功能活性原料产品,主要应用于化妆品、健康食品和生物医药等领域。2024年1月,和晨生物获得合肥市科技局的授牌,作为“合肥市合成生物学生物活性原料科技成果转化中试基地”,公司向高校、科研院所和相关企业开放服务,为其提供从实验室到产业化的中试验证。
9.金禾实业
安徽金禾实业股份有限公司(002597.SZ)位于滁州市来安县,是一家致力于高端甜味剂、食用香料细分领域的服务美好生活和先进制造的高新技术企业。主要产品有安赛蜜、麦芽酚、乙基麦芽酚、甲基环戊烯醇酮等,产品已覆盖食品、饮料、香精香料、健康保健品等领域,是国家“制造业单项冠军”企业。金禾实业积极布局合成生物赛道,合成生物产品涉及甜菊糖苷、罗汉果甜苷、阿洛酮糖、红没药醇、圆柚酮等食品添加剂和日化香料品类。公司持续关注非粮生物基材料全组分高效利用的创新技术,推动产品的生物催化和胞内合成路径的绿色生产,与高校、科研院所合作开发利用微生物发酵生产多款天然甜味剂和香料产品。
10.瑞邦生物
安徽瑞邦生物科技有限公司位于马鞍山市当涂经济开发区,是一家集科研、制造、国内外市场销售为一体的、以“医药健康、动物营养及健康护理”为主业的生物制造类国家高新技术企业,瑞邦生物为江西海文生物科技有限公司全资子公司。瑞邦生物采用合成生物技术生产的维生素B3类产品烟酰胺在国内市场占有率稳居行业首位,累计出口全球70多个国家和地区,荣膺国家级专精特新“小巨人”企业、“省制造业单项冠军培育企业”等称号,并有望获评国家级制造业单项冠军企业。公司积极与江南大学、南昌大学等建立产学研合作,致力打造烟酰胺/烟酸合成生物全产业链企业。
四、2025年安徽省合成生物投融资事件
2025年1-7月安徽合成生物产业相关投融资涉及生物医药、生物基材料与化学品、农业环保、生命科学工具等细分赛道,主要集中在合成生物下游产业链。合成生物学在医药、材料、生物基化学品等领域的加速商业化,同时地方国资深度参与,推动区域产业集群发展,合肥为投资热点地市。
表6 2025年安徽合成生物产业相关投融资事件
五、总结
安徽省合成生物产业已进入快速发展阶段,以合肥、蚌埠、芜湖为代表的产业集聚区初步形成,依托中国科大、中科院合肥物质研究院、合肥综合性国家科学中心大健康研究院等科研机构的技术优势,在基因编辑、代谢途径优化、工业发酵等领域取得显著突破,华恒生物突破大宗化学品的低成本生物合成、丰原集团推动生物基材料规模化生产、阿法纳生物和星眸生物等创新疗法研发初现。当前合肥市通过专项政策布局合成生物产业园促进产业集群发展,蚌埠市加快培育生物基新材料产业世界之都,芜湖市明确千亿级产业发展目标。省级未来产业先导区长丰县以“南研北产”模式深化产学研创新合作,全省“政产学研金服用”协同平台初具规模。未来随着合成生物技术与“双碳”战略深度融合,生物能源、CO₂转化等绿色技术将加速应用,推动产业集群向高端化、国际化跃升。然而,与北京、上海、广东、浙江、江苏等先发省份相比,安徽在合成生物产业整体规模、龙头企业带动效应以及资本活跃度等方面仍存在一定差距,建议进一步强化基础研究与产业转化的双向赋能,优化区域分工(合肥聚焦生物医药、生物基化学品与材料,蚌埠大力发展生物基新材料、芜湖侧重农业与食品),加快建设中试平台与生物铸造工厂等共性设施,设立专项天使基金支持技术成果转化应用,同时加快拓展碳足迹和碳认证等产业衍生服务,构建长三角协同创新网络,为安徽省合成生物产业可持续发展提供技术、资本与政策保障。
参考资料:
安徽工信 微信公众号《安徽:全方位布局生物制造 高质量发展再添“绿色动力”》
双凤发布 微信公众号《聚力打造省内合成生物产业发展新高地》
安徽日报 《聚力打造蚌埠“材料之都”新名片》
芜湖新闻网 《我市出台合成生物产业发展行动方案》
END
引进/卖出
100 项与 浙江工业大学 相关的药物交易
登录后查看更多信息
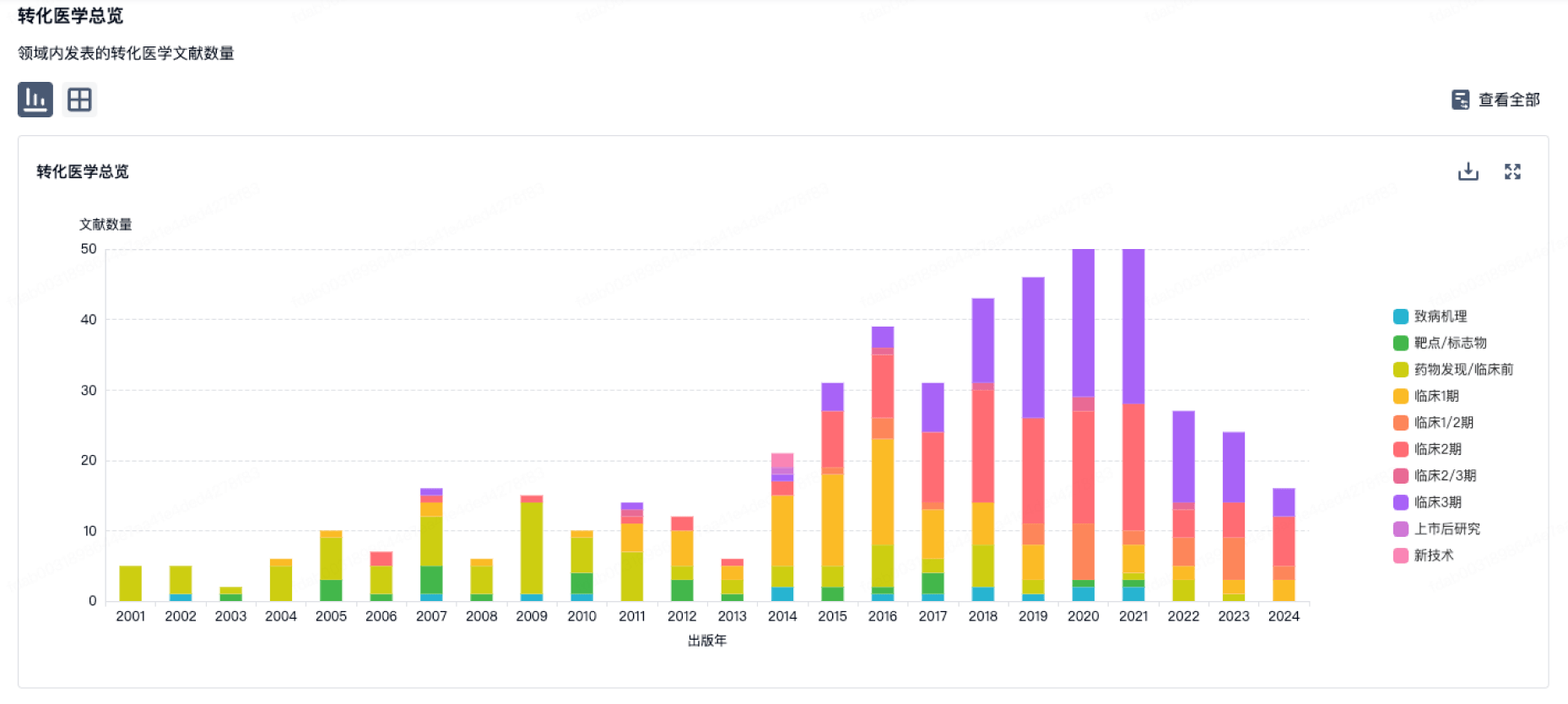
100 项与 浙江工业大学 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年10月20日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
药物发现
20
10
临床前
其他
1
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
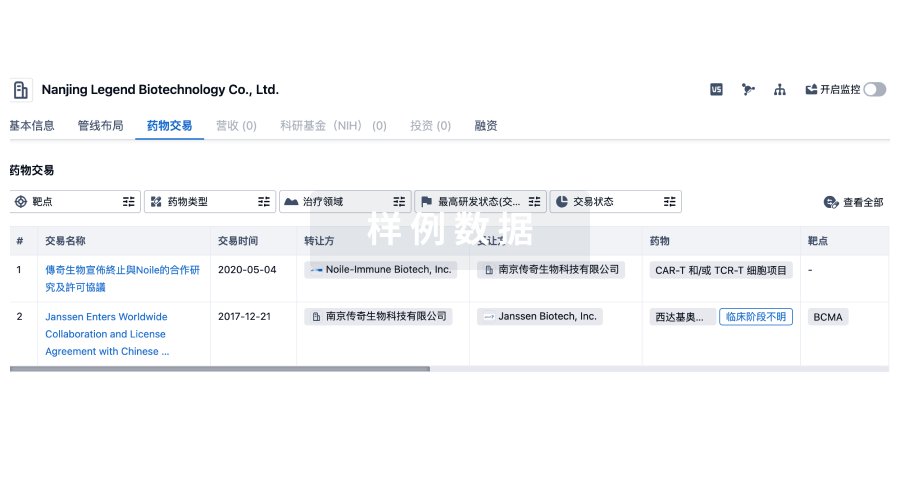
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
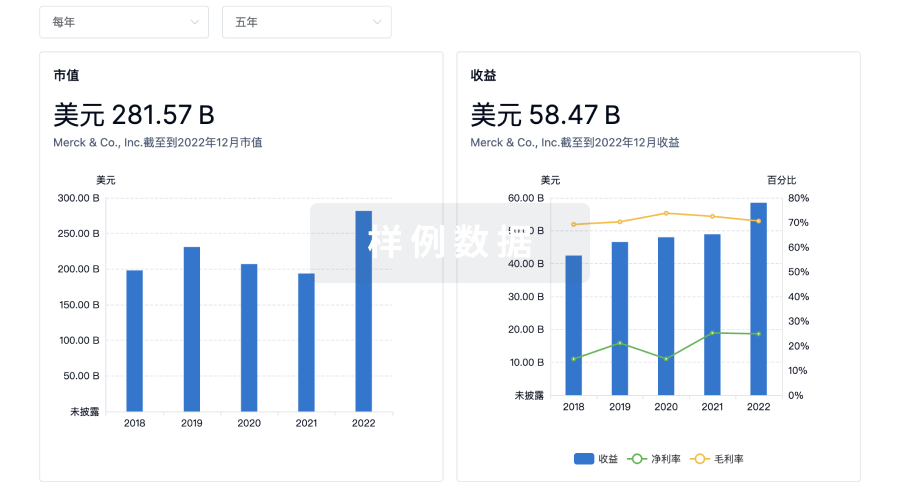
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
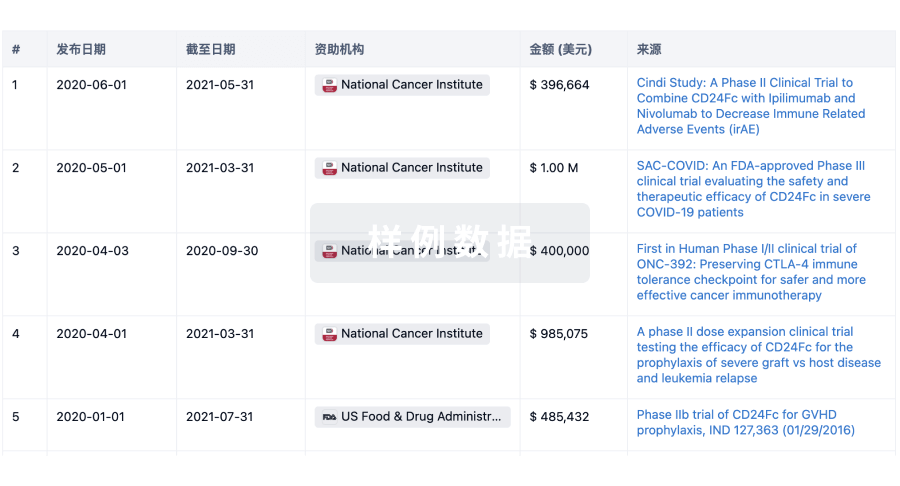
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

