预约演示
更新于:2025-05-07
Developmental Disabilities
发育障碍
更新于:2025-05-07
基本信息
别名 Child Development Deviation、Child Development Deviations、Child Development Disorder + [45] |
简介 Disorders in which there is a delay in development based on that expected for a given age level or stage of development. These impairments or disabilities originate before age 18, may be expected to continue indefinitely, and constitute a substantial impairment. Biological and nonbiological factors are involved in these disorders. (From American Psychiatric Glossary, 6th ed) |
关联
53
项与 发育障碍 相关的药物作用机制 叶酸受体调节剂 [+3] |
原研机构- |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区 韩国 |
首次获批日期2024-08-19 |
靶点 |
作用机制 GnRHR拮抗剂 |
在研机构 |
最高研发阶段批准上市 |
首次获批国家/地区 欧盟 [+3] |
首次获批日期2022-06-14 |
靶点 |
作用机制 PI3Kα抑制剂 |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2019-05-24 |
1,074
项与 发育障碍 相关的临床试验ACTRN12624001244594
Impact of CP-KASP (Cerebral Palsy Knowledge, Advocacy Skills, and Support Program) on parenting stress, empowerment and efficacy, and quality of life in caregivers of children with cerebral palsy.
开始日期2026-07-01 |
申办/合作机构 |
ACTRN12625000363482
Kids Connect Integrated Hybrid program: a tiered care approach to optimise access to screening and support for child’s developmental, parental mental health, and family psychosocial needs for ALL families (including priority populations).
开始日期2025-05-01 |
申办/合作机构- |
ACTRN12625000339459
Evaluating the safety and clinical effectiveness of the M-Finity stem in patients undergoing total hip arthroplasty through a CT-based migration analysis.
开始日期2025-04-30 |
申办/合作机构- |
100 项与 发育障碍 相关的临床结果
登录后查看更多信息
100 项与 发育障碍 相关的转化医学
登录后查看更多信息
0 项与 发育障碍 相关的专利(医药)
登录后查看更多信息
91,939
项与 发育障碍 相关的文献(医药)2026-01-01·Neural Regeneration Research
GEMIN5 and neurodevelopmental diseases: From functional insights to disease perception
Article
作者: Francisco-Velilla, Rosario ; Martinez-Salas, Encarnacion
2025-12-31·Future Science OA
Validation of salivary proteomic biomarkers for early detection of oral cancer in the Egyptian population
Article
作者: Ghalwash, Dalia ; Ammar, Ahmed ; Abou-Bakr, Asmaa ; El-Gawish, Ayman ; Diab, Al-Hassan
2025-12-31·Expert Review of Vaccines
Immunogenicity and safety of a live attenuated varicella vaccine in healthy subjects aged between 13 to 55 years: a double-blind, randomized, active-controlled phase III clinical trial in China
Article
作者: Sun, Jinfang ; Zhang, Hao ; Zhang, Yang ; Pan, Hongxing ; Li, Jingxin ; Zhu, Fengcai ; Shi, Jinhui ; Chang, Xianyun ; Li, Guifan ; Wang, Shiyuan
955
项与 发育障碍 相关的新闻(医药)2025-05-02
Highlights Continued Progress in Early Detection and the Progression of Esophageal Disease
SUFFERN, N.Y., May 2, 2025 /PRNewswire/ -- CDx Diagnostics announced today that new clinical data supporting the utility of the WATS3D test for Barrett's esophagus and dysplasia screening and surveillance has been selected for podium presentation at the 2025 Digestive Disease Week (DDW) in San Diego, the premier conference for gastroenterology professionals. This podium presentation, along with additional data featured in a separate poster presentation, highlights WATS3D's vital role in early detection, progression monitoring and risk assessment of patients at risk for Barrett's esophagus and esophageal cancer.
Continue Reading
WATS3D has consistently shown superior detection rates compared to conventional forceps biopsy (FB), the latter of which can miss up to 50% of cases. By sampling a broader area of mucosa and analyzing more cellular material using advanced 3D imaging, WATS3D may help gastroenterologists detect Barrett's esophagus that might otherwise go undiagnosed.
New DDW 2025 data reveal WATS3D's enhanced capabilities for Barrett's detection, surveillance and risk assessment.
Post this
Key Events at DDW 2025:
Podium Presentation: "Progression Rates of Barrett's Esophagus and Dysplasia Detected by WATS3D Are Similar to Those Diagnosed by Forceps Biopsies"
Poster Presentation: "MUC2 Immunostaining Predicts Neoplastic Progression as Accurately as the Presence of Goblet Cells in Columnar-Lined Esophagus: A WATS3D Study of 3,553 Patients"
Nicholas J. Shaheen, MD will deliver the podium presentation, based on a collaborative study with Dr. Michael Smith, Dr. Vivek Kaul, Mendel Singer Ph.D., and Dr. Robert Odze.
"The findings validate WATS3D's role in early detection," said Dr. Shaheen. "We're excited to share these results at DDW and discuss their impact on patient care."
Robert D. Odze, MD noted that "These studies advance our understanding of the biological significance of WATS3D detected lesions and their risk of progression to cancer in Barrett's esophagus."
The new data shared at DDW 2025 from May 3-6 offers important clinical insights into the evolving role of WATS3D in Barrett's detection, surveillance and risk assessment. For more information, visit or contact us at [email protected].
About CDx Diagnostics
CDx Diagnostics' mission is to Empower Physicians with Innovative Technology to Prevent Esophageal Cancer, One Patient at a Time. The company's proprietary diagnostic platform combines advanced computer imaging, artificial intelligence, molecular biology, and 3D cytopathology to detect precancerous changes earlier and more precisely than conventional methods. This pioneering solution has analyzed over 400,000 cases, empowering providers to identify those requiring early intervention, reduce time to treatment, and improve outcomes by detecting disease that might be missed by traditional forceps biopsies.
CONTACT:
Logan Garrett
[email protected]
423-519-9979
SOURCE CDx Diagnostics
WANT YOUR COMPANY'S NEWS FEATURED ON PRNEWSWIRE.COM?
440k+
Newsrooms &
Influencers
9k+
Digital Media
Outlets
270k+
Journalists
Opted In
GET STARTED
诊断试剂
2025-05-01
·梅斯医学
6天大的小雅,出生时体重仅2.3公斤,是个看起来略显瘦弱的新生儿。刚出生几天,她就出现了反复哭闹、吃奶差、尿多等情况。起初家人以为是“肠胀气”或是“没吃饱”,并未太在意。但很快,小雅的情况急转直下——面色发灰、反应迟钝、体重不增,还出现了明显的脱水迹象。家人慌忙将她送至医院急诊。血糖检查结果让人震惊:空腹血糖高达19 mmol/L,明确诊断为新生儿糖尿病(NDM)。进一步检查显示,小雅无酮症酸中毒,但需立即启动胰岛素治疗以稳定血糖和纠正脱水。医生随即安排了基因检测,结果发现小雅携带KCNJ11基因突变,明确为KATP通道缺陷导致的NDM。分型新生儿糖尿病是一组异质性单基因遗传病,临床少见,多发生在6月龄以内,少数发病年龄可达12月龄。按病程NDM可分为暂时性新生儿糖尿病(TNDM),永久性新生儿糖尿病(PNDM)和NDM相关综合征。NDM罕见,新生儿糖尿病的发生率为1/50万~1/40万,我国尚无相关数据报道。其中约50%是PNDM,50%是TNDM。临床表现新生儿糖尿病(Neonatal Diabetes Mellitus, NDM)是一种罕见的单基因病,其共同临床特征包括宫内发育迟缓、低出生体重、发育不良、多尿和严重脱水,部分患儿还可伴有出生缺陷、肌无力和神经系统异常。TNDM通常表现为出生后1周内出现严重的非酮症性高血糖,12周内可自行缓解,但约50%~60%的患者在青春期前后复发,复发时症状类似2型糖尿病。PNDM通常表现为出生时小于胎龄,糖尿病为唯一临床表现,但某些基因变异可伴胰腺外表现,如NEUROD1突变常合并中枢神经异常,HNF1β突变则可见肾脏或生殖系统异常。PNDM无自然缓解期。NDM还可能为多种综合征的一部分,如Wolcott-Rallison综合征、IPEX综合征和Mitchell-Riley综合征等,表现更为复杂,需综合评估。诊断新生儿糖尿病(NDM)的诊断依赖于临床表现、实验室检查及基因检测,其中基因检测是进行分型诊断的金标准。典型诊断标准包括新生儿期或生后6周内出现持续空腹血糖≥7 mmol/L(120 mg/dL),病程超过2周,且需依赖胰岛素治疗以维持血糖正常水平。确诊及具体分型需通过基因检测,传统方法主要为Sanger测序。鉴别诊断应包括1型糖尿病、早发型2型糖尿病及其他单基因糖尿病(如MODY)。其中,1型糖尿病抗体检测及相关基因检测对于明确病因具有重要意义。对于疑似NDM的患儿,应遵循规范的诊断流程,以避免误诊和漏诊。治疗治疗原则包括纠正脱水和酸中毒、胰岛素替代、感染控制及家属教育。液体治疗分阶段进行,并根据脱水程度调整。所有患儿应先接受胰岛素治疗,酮症酸中毒者推荐静脉持续滴注;无酮症者可使用胰岛素泵。在确诊为KATP通道基因突变后,可逐步转换为磺酰脲类药物治疗。参考资料:[1]王冬梅,程明,曹冰燕.新生儿糖尿病的研究进展[J].临床内科杂志,2024,41(08):519-522.[2] 中华医学会儿科学分会内分泌遗传代谢学组. 儿童单基因糖尿病临床诊断与治疗专家共识[J]. 中华儿科杂志,2019,57(7):508-514.[3] Hattersley Andrew T,Greeley Siri A W,Polak Michel et al. ISPAD Clinical Practice Consensus Guidelines 2018: The diagnosis and management of monogenic diabetes in children and adolescents.[J] .Pediatr Diabetes, 2018, null: 47-63.来源 | 梅斯医学编辑 | wanny神经系统罕见病交流群↓点击下方“阅读原文”,下载梅斯医学APP吧!
临床研究申请上市
2025-04-28
PEABODY, Mass., April 28, 2025 /PRNewswire/ -- Privo Technologies, Inc., a clinical-stage biopharmaceutical company pioneering nanotechnology-based cancer therapies, is thrilled to announce the activation of multiple premier clinical sites across the United States for its ongoing CLN-004 trial. This Phase 2/3, open-label, two-arm study is evaluating the safety and efficacy of two of Privo's lead assets—PRV111 and PRV211—in patients with head and neck squamous cell carcinoma (HNSCC).
The trial is now actively recruiting at the following esteemed institutions:
University of Chicago Medicine (Chicago, IL)
City of Hope (Los Angeles, CA)
Miami Cancer Institute (Miami, FL)
Cleveland Clinic (Cleveland, OH)
Additional leading cancer centers are in the process of being activated and will be announced in the coming weeks.
"We are honored to collaborate with some of the nation's leading head and neck oncology experts at these top-tier institutions," said Dr. Manijeh Goldberg, CEO of Privo Technologies. "Their participation underscores the potential impact of our novel therapies in transforming the treatment landscape for patients with HNSCC."
About the CLN-004 Trial
The CLN-004 study comprises two distinct arms:
Arm 1: Evaluates PRV111, a nanoengineered, self-adhesive patch designed for the topical delivery of cisplatin directly to the tumor site. This arm targets patients with carcinoma in situ (CIS) and high-grade dysplasia of the oral cavity, aiming to improve local control and potentially reduce the extent of surgical intervention.
Arm 2: Assesses PRV211, an intraoperative chemotherapy system intended for application into the resected tumor bed during surgery. This approach seeks to eliminate residual microscopic disease and reduce the risk of tumor recurrence in patients with stages T1-T4 HNSCC.
Both PRV111 and PRV211 are part of Privo's proprietary PRV™ platform, which utilizes nanotechnology to enhance the locoregional delivery of chemotherapeutic agents, aiming to maximize therapeutic efficacy while minimizing systemic toxicity.
About Privo Technologies
Privo Technologies is dedicated to developing innovative therapies that address unmet needs in oncology, with a focus on mucosal cancers. By leveraging its PRV™ platform, Privo aims to improve patient outcomes through targeted, localized drug delivery systems.
For more information about the CLN-004 clinical trial or to inquire about participation, please visit
.
Media Contact:
Stefanie Cantin
Director of Communications
Privo Technologies, Inc.
Email: [email protected]
Phone: (978) 587-2322
Note: This press release is based on information available as of April 28, 2025. For the most current details regarding the CLN-004 trial and participating sites, please refer to Privo Technologies' official website or contact the company directly.
SOURCE Privo Technologies
WANT YOUR COMPANY'S NEWS FEATURED ON PRNEWSWIRE.COM?
440k+
Newsrooms &
Influencers
9k+
Digital Media
Outlets
270k+
Journalists
Opted In
GET STARTED
临床研究ASCO会议
分析
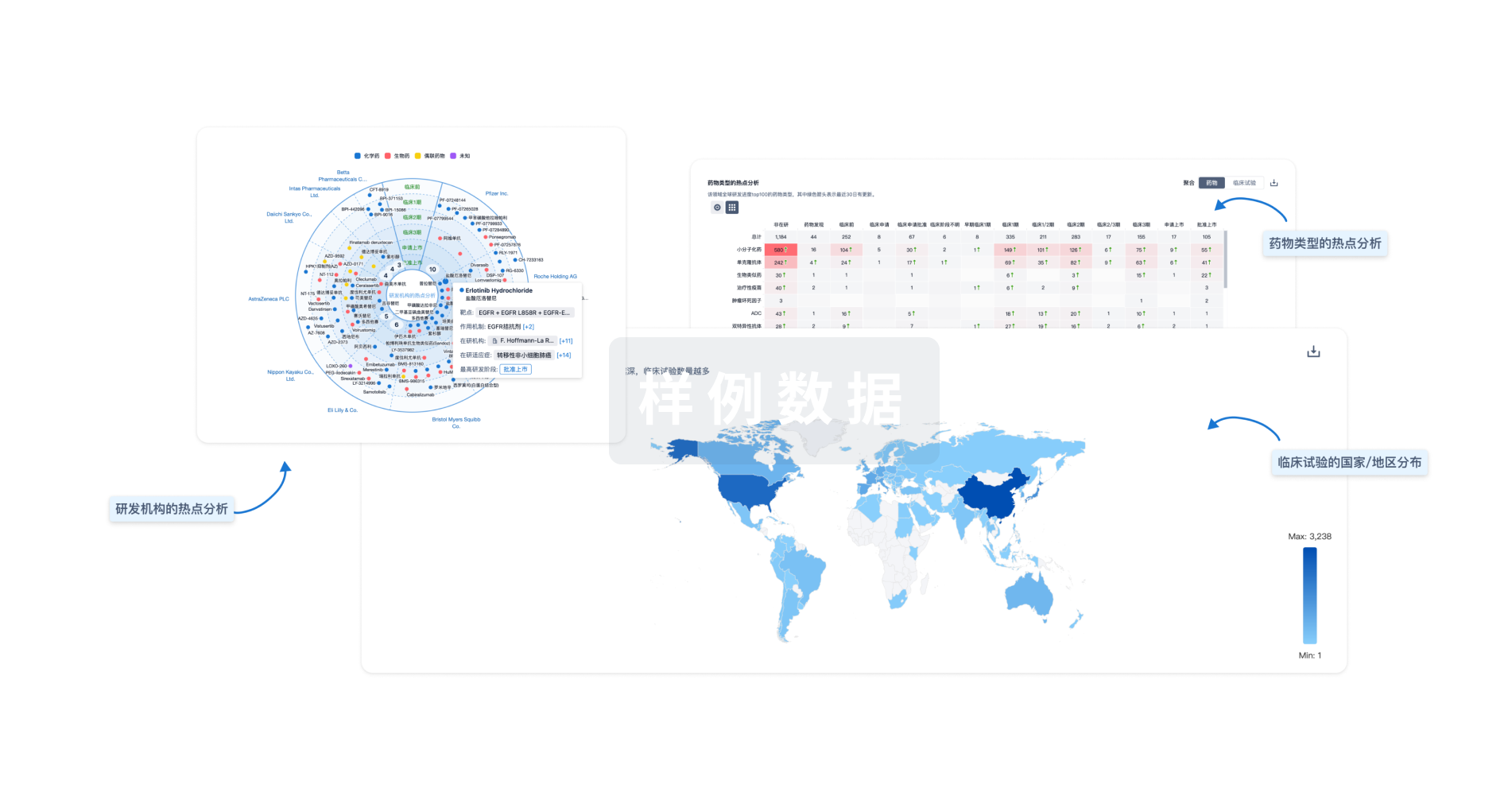
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用