预约演示
更新于:2025-05-07
Primary Parkinson's disease
原发性帕金森病
更新于:2025-05-07
基本信息
别名- |
简介- |
关联
16
项与 原发性帕金森病 相关的药物靶点 |
作用机制 COMT抑制剂 |
非在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 欧盟 [+3] |
首次获批日期2016-06-24 |
作用机制 5-HT2A receptor反向激动剂 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2016-04-29 |
靶点 |
作用机制 A2aR拮抗剂 |
非在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 日本 |
首次获批日期2013-03-25 |
86
项与 原发性帕金森病 相关的临床试验CTR20250991
甲磺酸溴隐亭片在健康受试者中随机、开放、单剂量、四周期、两序列、完全重复交叉的餐后状态下生物等效性试验
主要研究目的
研究餐后状态下单次口服受试制剂甲磺酸溴隐亭片(规格:2.5mg,浙江泰利森药业有限公司)与参比制剂甲磺酸溴隐亭片(商品名:Parlodel®,规格:2.5mg;DOPPEL Farmaceutici S.r.l.公司生产)在健康成年受试者体内的药代动力学,评价餐后状态口服两种制剂的生物等效性。
次要研究目的
研究受试制剂甲磺酸溴隐亭片和参比制剂甲磺酸溴隐亭片(Parlodel®)在健康成年受试者中的安全性。
开始日期2025-04-13 |
申办/合作机构 |
NCT06888869
The Development and Verification of an Interactive Home-based Rehabilitation Exercise Assessment Platform and Exploration of Technology Acceptance and Clinical Effects in Patients with Parkinson's Disease
This study aims to develop an "Interactive Home-based Rehabilitation Exercise Assessment Platform" that incorporates visual feedback similar to virtual reality into rehabilitation machines and aerobic cycling programs. In the clinical research part, 92 Parkinson's disease patients will be randomly assigned to three groups: the clinical rehabilitation group, the home-based rehabilitation group, and the control group. Each group will undergo the intervention twice a week, with each session lasting about 30-45 minutes over a period of 12 weeks for a total of 24 combined resistance and aerobic rehabilitation training sessions. The effectiveness evaluation will include measurements of upper limb grip strength, lower limb muscle strength, the 3-meter sit-to-stand test, the 6-minute walk test, the Parkinson's Disease Severity Scale, the Quality of Life Scale, the 10-meter walk test, and technology acceptance forms.
开始日期2025-03-21 |
申办/合作机构 |
CTR20244974
UX-DA001注射液(人中脑多巴胺能神经前体细胞注射液)在原发性帕金森病患者中的探索性临床试验
主要目的:评估不同剂量组 UX-DA001 注射液细胞移植治疗原发性帕金森病(PD)患者的安全性和耐受性。
次要目的:
评估不同剂量组 UX-DA001 注射液在原发性 PD 患者脑内的植入、存活和增生情况;
评估不同剂量组 UX-DA001 注射液对原发性 PD 患者运动症状的改善情况;
评估不同剂量组 UX-DA001 注射液对原发性 PD 患者非运动症状及整体生活质量的影响。
开始日期2025-02-21 |
申办/合作机构 |
100 项与 原发性帕金森病 相关的临床结果
登录后查看更多信息
100 项与 原发性帕金森病 相关的转化医学
登录后查看更多信息
0 项与 原发性帕金森病 相关的专利(医药)
登录后查看更多信息
355
项与 原发性帕金森病 相关的文献(医药)2026-01-01·Neural Regeneration Research
Adeno-associated viral vectors for modeling Parkinson’s disease in non-human primates
Article
作者: Lanciego, José L. ; Chocarro, Julia
2024-12-01·Clinical Genitourinary Cancer
Appearance of New Lesions Associate With Poor Prognosis in Pembrolizumab-Treated Urothelial Carcinoma
Article
作者: Teishima, Jun ; Okamura, Yasuyoshi ; Nakano, Yuzo ; Bando, Yukari ; Hyodo, Yoji ; Hara, Takuto ; Terakawa, Tomoaki ; Suzuki, Kotaro ; Chiba, Koji ; Miyake, Hideaki
2024-10-30·Brain Communications
Brain age in genetic and idiopathic Parkinson's disease
Article
作者: Storch, Alexander ; Schumacher, Julia ; Hermann, Andreas ; Hoffmann, Hauke ; Dyrba, Martin ; Teipel, Stefan J
42
项与 原发性帕金森病 相关的新闻(医药)2025-04-12
·医药观澜
▎药明康德内容团队报道根据中国国家药监局药品审评中心(CDE)官网以及公开资料梳理,本周(4月7日~4月12日),有16款1类创新药首次在中国获得临床试验默示许可(IND)。这些产品包括小分子抑制剂、小分子蛋白降解剂、放射性治疗药物、双抗、抗体偶联药物(ADC)、多肽偶联药物(PDC)、siRNA药物、CAR-T疗法、iPSC衍生细胞疗法等类型。田边三菱制药:MT-7117片作用机制:MC1R选择性激动剂适应症:红细胞生成性原卟啉病/X连锁原卟啉病田边三菱制药(Mitsubishi Tanabe Pharma)申报的1类新药MT-7117片获批临床,拟开发治疗红细胞生成性原卟啉病(EPP)/X连锁原卟啉病(XLP)。公开资料显示,MT-7117(通用名:dersimelagon)是一种新型合成、口服给药小分子药物,为黑皮质素-1受体(MC1R)的选择性激动剂。EPP和XLP均属于红细胞生成性卟啉症(EPs),这是一种可能危及生病的罕见疾病,由影响血红素合成通路的突变引起,其特点是严重的皮肤敏感性,表现为剧烈的灼痛发作,伴有持续的水泡、水肿和皮肤损毁。从作用机制上看,MC1R激动剂能够激活黑素细胞并刺激真黑素(eumelanin)的合成。真黑素通过吸收和散射紫外线,清除自由基和活性氧,并作为一种中性密度过滤器,能够减少所有波长的光的透射,从而帮助抵抗紫外线。云核医药:INR102注射液作用机制:靶向PSMA的放射性治疗药物适应症:PSMA阳性mCRPC云南白药全资子公司云核医药申报的1类新药INR102注射液获批临床,拟用于治疗已经接受过雄激素受体通路抑制剂和紫杉烷类药物化疗的PSMA阳性的转移性去势抵抗性前列腺癌(mCRPC)成人患者。根据云南白药公告介绍,这是云核医药研发的化学1类放射性治疗类创新药。该产品活性分子结构包含靶向配体、放射性同位素镥[177Lu],靶向配体可特异性结合PSMA表达阳性的癌细胞,携带的镥[177Lu]衰变过程中可发射β射线,导致癌细胞DNA损伤,最终诱导细胞死亡。艾奇西医药、中国医学科学院医药生物技术研究所、赛隆药业:注射用IMBZ18g作用机制:抗革兰氏阴性耐药菌机制适应症:复杂性尿路感染(包括急性肾盂肾炎)公开资料显示,IMBZ18g是一款拥有全新的化合物结构和抗革兰氏阴性耐药菌机制的1类新药,有着较好的耐药菌活性,有望为治疗中重度阴性菌感染的患者提供更安全、有效的治疗新选择。中国科学院上海药物研究所、烟台新药创制山东省实验室、中科中山药物创新研究院、浙江大学:TT-9701片作用机制:CHK1小分子口服抑制剂适应症:复发难治性急性髓系白血病公开资料显示,TT-9701片(开发代号:TT-97)是由中科中山药物创新研究院李佳团队、浙江大学刘滔团队联合研发的一款高选择性CHK1小分子口服抑制剂,TT-97在体外对急性髓系白血病细胞敏感,在体内移植瘤和血液瘤模型上,TT-97能抑制多种急性髓系白血病肿瘤模型的肿瘤增长,以及显著延长白血病模型小鼠的生存期,且安全可控。福元医药:FY101注射液作用机制:化药1类新药适应症:高脂血症根据福元医药公告,FY101注射液为化学药品1类创新药,适用于高脂血症的治疗,给药途径为皮下注射。目前国际范围内尚无同类型同靶点产品获批上市。恒瑞医药:SHR-4658注射液作用机制:生物制品新药适应症:心力衰竭恒瑞医药1类新药SHR-4658注射液获批临床,拟开发治疗心力衰竭。目前尚未从公开渠道查询到该产品的具体作用机制,从受理号可知这是一款生物制品新药。冰洲石生物:AC699片作用机制:ERα嵌合降解剂适应症:ER+/HER2-乳腺癌冰洲石生物科技(Accutar Biotechnology)申报的1类新药AC699片获批临床,拟用于治疗ESR1突变的雌激素受体阳性(ER+)和人表皮生长因子受体2阴性(HER2-)晚期或转移性乳腺癌。公开资料显示,AC699是一种口服生物可利用的ERα嵌合降解剂,它通过将ER配体有效地连接到E3连接酶招募配体上,使ERα靠近E3连接酶,导致ERα的泛素化和随后的降解。E3连接酶嵌合降解代表了一项技术进步,有潜力诱导有效和更深的ER降解。正大天晴:TQB3019胶囊作用机制:BTK蛋白降解靶向嵌合体适应症:血液肿瘤TQB3019是一款口服、靶向BTK的蛋白降解靶向嵌合体(PROTAC) 药物。根据正大天晴新闻稿介绍,作为一种异双功能分子,TQB3019通过结合BTK蛋白和E3泛素连接酶,启动泛素蛋白酶体途径,降解靶蛋白。该作用机制可以抑制BTK及下游信号传导,进而抑制肿瘤生长。与传统的BTK抑制剂相比,PROTAC药物在攻克BTK耐药难题上展现出更大潜力。体外研究显示,TQB3019对多种体外培养BTK野生型及不同突变类型的肿瘤细胞均有显著的增殖抑制作用。在动物模型中,TQB3019可以显著抑制BTK野生型及不同耐药突变细胞的小鼠皮下移植瘤的生长,且抑制活性强。先声药业:注射用SIM0686作用机制:靶向FGFR2b的ADC适应症:FGFR2b阳性的局部晚期/转移性实体瘤SIM0686为先声药业旗下抗肿瘤创新药公司先声再明开发的ADC项目。其临床前数据已入选2025年美国癌症研究协会年会(AACR)。该分子结合了抗体分子的肿瘤特异靶向性,以及拓扑异构酶抑制剂的肿瘤杀伤性,在多项临床前研究中显示出显著抗肿瘤疗效,不仅可抑制FGFR2b表达阳性的肿瘤细胞的增殖,还可通过旁观者效应对FGFR2b表达阴性的肿瘤细胞起到杀伤效果。本次获批临床后,先声再明即将启动SIM0686的1期临床研究,以评估其在晚期实体瘤患者中的安全性、耐受性、药代动力学、药效学及初步抗肿瘤活性。士泽生物:XS411细胞注射液作用机制:iPSC衍生多巴胺能神经前体细胞注射液适应症:原发性帕金森病XS411是士泽生物开发的异体通用“现货型”iPSC衍生多巴胺能神经前体细胞注射液。根据士泽生物新闻稿介绍,在此前该公司与合作医院开展的国家级备案临床研究中,临床级iPSC衍生细胞经患者纹状体壳核区移植治疗中重度帕金森病最长随访期已超过12个月,无细胞疗法相关不良事件出现,且多例受试患者开关期时间及MDS-UPDRS评分量表等关键疗效指标及多项非运动指标均获得显著性改善。天士力:P134细胞注射液作用机制:靶向CD44和(或)CD133的自体CAR-T产品适应症:复发胶质母细胞瘤天士力1类新药P134细胞注射液获批临床,拟开发治疗复发胶质母细胞瘤。根据天士力公告,这是是由天士力与北京神经外科研究所共同合作开发的创新生物药,是一款靶向CD44和(或)CD133的自体CAR-T产品。该产品的作用机制为特异性识别并结合在原发性和复发性胶质母细胞瘤(GBM)中呈现特异性互斥高表达的抗原靶标,高效激活并延长T细胞寿命,从而杀伤肿瘤细胞。大睿生物:RN1871注射液作用机制:化药适应症:原发性高血压大睿生物1类新药RN1871注射液获批临床,拟开发治疗原发性高血压。目前尚未从公开渠道查询到该产品的具体作用机制。公开资料显示,大睿生物是一家核酸药物平台公司,致力于发现和开发创新性的核酸药。鼎新基因:DNV001注射液作用机制:PCSK9靶向siRNA药物适应症:血脂异常鼎新基因旗下鼎乐新为研发的1类新药DNV001注射液获批临床,拟用于血脂异常的治疗,包括以低密度脂蛋白胆固醇(LDL-C)升高为特征的成人原发性高胆固醇血症(杂合子型家族性和非家族性)或混合型血脂异常患者的治疗。根据鼎乐新为公开信息,DNV001是一种GalNAc-siRNA,通过敲降肝脏细胞中PCSK9 mRNA,从源头上阻断PCSK9的表达。药学和临床前研究表明,该产品具有很好的稳定性、安全性和有效性,有望实现临床一年仅两次注射达到长效降脂的目标。石药集团:SYH2046片作用机制:小分子药物适应症:急性心肌梗死后心力衰竭根据石药集团公告,SYH2046片是一款全新结构的小分子药物,本次获批临床的适应症为急性心肌梗死后心力衰竭。临床前研究表明,该产品可显著改善疾病动物模型的心脏功能并降低心脏不良重构,且具有较高的安全性。与传统心衰药物相比,该产品具有全新作用机制,有望通过改善心脏细胞的代谢,在心肌损伤早期积极促进组织修复,以恢复心肌梗死后的心脏功能。康方生物:AK139注射液作用机制:靶向IL-4Rα/ST2的双抗适应症:COPD、支气管哮喘康方生物1类新药AK139注射液获批临床,拟开发治疗控制欠佳的慢性阻塞性肺疾病(COPD)、中重度控制欠佳的支气管哮喘。公开资料显示,这是康方生物研发的靶向IL-4Rα/ST2双特异性抗体。该产品可同时阻断IL-4、IL13(通过结合IL-4和IL-13受体复合物共享的IL-4Rα亚单位)和IL-33/ST2介导的炎症通路。在临床前研究中,该产品在抑制炎症因子释放、组织炎症细胞浸润等指标中展现出显著优于IL-4或ST2单靶点抗体的协同效应。贝海生物:注射用BH259作用机制:多肽偶联药物(PDC)适应症:晚期实体瘤根据贝海生物公开资料,BH259是一款全新的多肽偶联药物。在多项临床前研究中,BH259表现出优异的抗肿瘤活性和安全性,在动物体内药效试验结果显示,BH259在具有良好抗肿瘤活性同时能够显著增大了payload毒素的安全性及治疗窗口。在药代动力学研究中,BH259能够成功靶向肿瘤细胞及组织。参考资料:[1]中国国家药监局药品审评中心(CDE)官网. Retrieved Apr 5, From https://www.cde.org.cn/main/xxgk/listpage/4b5255eb0a84820cef4ca3e8b6bbe20c[2]各公司官网及公开资料本文来自药明康德内容团队,欢迎个人转发至朋友圈,谢绝媒体或机构未经授权以任何形式转载至其他平台。转载授权或其他合作需求,请联系wuxi_media@wuxiapptec.com。免责声明:药明康德内容团队专注介绍全球生物医药健康研究进展。本文仅作信息交流之目的,文中观点不代表药明康德立场,亦不代表药明康德支持或反对文中观点。本文也不是治疗方案推荐。如需获得治疗方案指导,请前往正规医院就诊。
抗体药物偶联物临床申请细胞疗法放射疗法免疫疗法
2025-04-10
·氨基观察
氨基观察-创新药组原创出品作者 | 黄恺血友病治疗进入新时代。4月10日,据NMPA官网,信念医药基因疗法波哌达可基获批上市,用于治疗中重度血友病B(先天性凝血因子IX缺乏症)成年患者。该药物是国内首款血友病基因疗法。联影医疗布局大模型。4月9日,联影发布“元智”医疗大模型,并同步推出覆盖影像诊断、临床治疗、医学科教、医院管理、患者服务等多场景的10余款医疗智能体。其中,基于数千万级医疗影像数据和数十万级医疗级精细标注数据训练打造的“元智”医疗影像大模型能支持10+影像模态、300种影像处理任务,在处理诸如复杂病灶诊断,器官分割等关键任务上,模型的精准度测评已超过95%。在过去的一天里,国内外医药市场还有哪些热点值得关注?让氨基君带你一探究竟。/ 01 /市场速递1)和铂医药宣布任命陈侑晨为首席财务官4月10日,和铂医药宣布,任命陈侑晨(YC)为首席财务官。YC将常驻中国上海和香港,直接向和铂医药创始人、董事长兼首席执行官王劲松博士汇报。/ 02 /医药动态1)天士力P134细胞注射液获临床许可4月10日,据CDE官网,天士力P134细胞注射液获临床许可,拟开展治疗复发胶质母细胞瘤的研究。2)合源生物纳基奥仑赛注射液获临床许可4月10日,据CDE官网,合源生物纳基奥仑赛注射液获临床许可,拟用于治疗至少3线治疗失败的自身免疫性溶血性贫血的研究。3)士泽生物XS411细胞注射液获临床许可4月10日,据CDE官网,士泽生物XS411细胞注射液获临床许可,拟开展治疗原发性帕金森病(PD)的研究。4)大睿生物RN1871注射液获临床许可4月10日,据CDE官网,大睿生物RN1871注射液获临床许可,拟开展治疗原发性高血压的研究。5)翊石医药SYH2046片获临床许可4月10日,据CDE官网,翊石医药SYH2046片获临床许可,拟用于急性心肌梗死后心力衰竭。6)知微生物TM471-1胶囊获临床许可4月10日,据CDE官网,知微生物TM471-1胶囊获临床许可,拟开展治疗慢性自发性荨麻疹的研究。7)宜联生物BNT326获临床许可4月10日,据CDE官网,宜联生物BNT326获临床许可,拟开展治疗晚期非小细胞肺癌的研究。8)康方生物AK139注射液获临床许可4月10日,据CDE官网,康方生物AK139注射液获临床许可,拟开展治疗中重度控制欠佳的支气管哮喘、控制欠佳的慢性阻塞性肺疾病的多项研究。9)贝海生物注射用BH259获临床许可4月10日,据CDE官网,贝海生物注射用BH259获临床许可,拟用于治疗晚期实体瘤。10)首款国产血友病基因疗法获批上市4月10日,据NMPA官网,信念医药基因疗法波哌达可基获批上市,用于治疗中重度血友病B(先天性凝血因子IX缺乏症)成年患者。该药物是国内首款血友病基因疗法。/ 03 /器械跟踪1)锦波生物“注射用重组Ⅲ型人源化胶原蛋白凝胶”获准日前,山西锦波生物“注射用重组Ⅲ型人源化胶原蛋白凝胶”获NMPA批准上市,适用于注射到中面部皮下至骨膜上层,以矫正中面部容量缺失和/或中面部轮廓缺陷。/ 04 /数字医疗日报1)联影发布“元智”医疗大模型4月9日,联影发布“元智”医疗大模型,并同步推出覆盖影像诊断、临床治疗、医学科教、医院管理、患者服务等多场景的10余款医疗智能体。PS:欢迎扫描下方二维码,添加氨基君微信号交流。
基因疗法上市批准临床申请
2025-04-10
·医药地理
4月10日,国家药品监督管理局药品审评中心(CDE)公示信息显示,士泽生物自主研发的1类新药XS411细胞注射液正式获批临床试验(IND),用于治疗原发性帕金森病。这不仅是该产品在国内首次获得IND许可,更标志着我国在iPS(诱导多能干细胞)衍生细胞治疗神经系统疾病领域实现重大突破。XS411作为通用型iPS衍生多巴胺能神经前体细胞产品,其技术架构具有三重革新意义:1)采用国际领先的iPS重编程技术,规避胚胎干细胞伦理争议,实现神经细胞的可控分化;2)通过HLA配型优化和免疫调节技术,突破传统自体细胞治疗周期长、成本高的局限;3)纹状体壳核区立体定向移植技术确保新生多巴胺能神经元精准整合至神经环路。在已完成的备案临床研究中,XS411展现出显著的临床价值:最长12个月随访期内未报告细胞治疗相关严重不良事件(SAE);关键运动功能评估量表(MDS-UPDRS)评分降低达40%,"开关期"异常波动时间缩短60%;认知障碍、睡眠质量等次要终点同步改善,揭示多维度治疗效应。该产品此前已通过国家卫健委/药监局"双备案",成为国内首个获批开展iPS衍生细胞治疗神经系统疾病的国家级临床研究项目。此次IND获批意味着研究范围从研究者发起的临床探索(IIT)正式升级为注册临床路径。帕金森病作为仅次于阿尔茨海默病的第二大神经退行性疾病,其治疗面临双重挑战:我国65岁以上人群发病率达1.7%,预计2030年患者总数突破500万;左旋多巴等药物仅能缓解症状,DBS手术存在感染风险,均无法逆转神经元退变。XS411通过直接补充缺失的多巴胺能神经元,从病理根源实现神经功能重建。临床前研究显示,移植细胞可在宿主脑内长期存活(>18个月),并建立功能性神经突触连接。根据研发进度预测,该产品有望在2026年完成II期临床,2028年提交NDA申请。END如需获取更多数据洞察信息或公众号内容合作,请联系医药地理小助手微信号:pharmadl001
临床申请细胞疗法临床2期临床1期
分析
对领域进行一次全面的分析。
登录
或
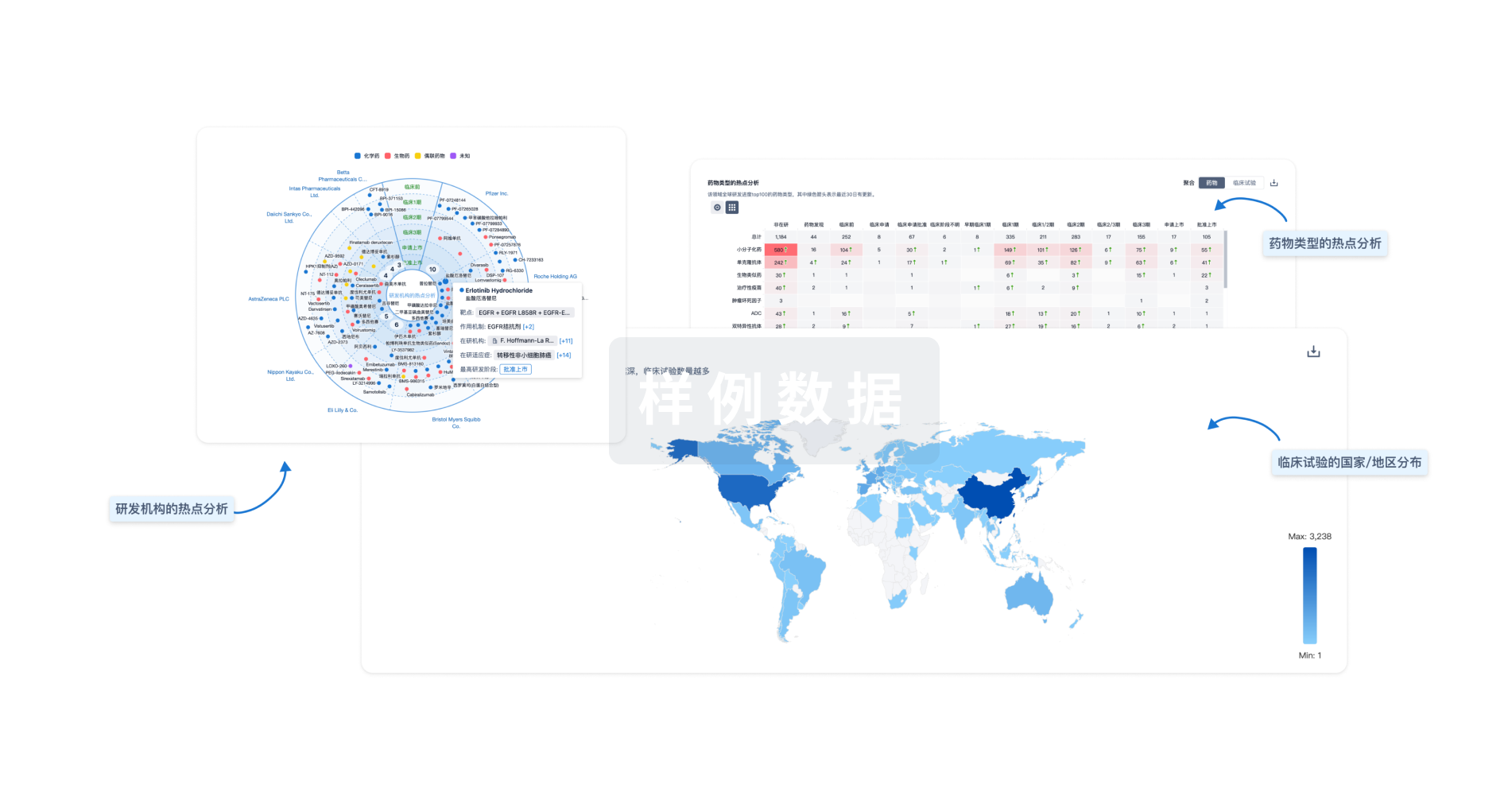
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用





