预约演示
更新于:2025-05-07
Camptothecin
喜树碱
更新于:2025-05-07
概要
基本信息
原研机构 |
在研机构 |
非在研机构- |
权益机构- |
最高研发阶段批准上市 |
首次获批日期 中国 (1982-01-01), |
最高研发阶段(中国)批准上市 |
特殊审评- |
登录后查看时间轴
结构/序列
分子式C20H16N2O4 |
InChIKeyVSJKWCGYPAHWDS-FQEVSTJZSA-N |
CAS号7689-03-4 |
关联
2
项与 喜树碱 相关的临床试验ACTRN12609000585224
A randomised phase II trial comparing the effect of Camptothecin-11 (CPT-11), Fluorouracil (5FU) and Folinic Acid versus CPT-11, 5FU and Folinic Acid followed by Oxaliplatin, 5FU and Folinic Acid on objective response rate in patients with metastatic colorectal cancer
开始日期2001-11-08 |
申办/合作机构- |
NCT00005820
A Phase II Study of 9 Nitrocamptothecin for Hormone Refractory Prostate Cancer
RATIONALE: Drugs used in chemotherapy use different ways to stop tumor cells from dividing so they stop growing or die.
PURPOSE: Phase II trial to study the effectiveness of nitrocamptothecin in treating men who have stage IV prostate cancer that has not responded to hormone therapy.
PURPOSE: Phase II trial to study the effectiveness of nitrocamptothecin in treating men who have stage IV prostate cancer that has not responded to hormone therapy.
开始日期2000-05-01 |
申办/合作机构 |
100 项与 喜树碱 相关的临床结果
登录后查看更多信息
100 项与 喜树碱 相关的转化医学
登录后查看更多信息
100 项与 喜树碱 相关的专利(医药)
登录后查看更多信息
10,157
项与 喜树碱 相关的文献(医药)2025-09-01·Biomaterials
Acid and phosphatase-triggered release and trapping of a prodrug on cancer cell enhance its chemotherapy
Article
作者: Shen, Zixiu ; Zhan, Wenjun ; Zhu, Liangxi ; Zhang, Ziyi ; Zhou, Lei ; Wang, Jue ; Wang, Rui ; Liu, Xiaoyang ; Zhao, Furong ; Tang, Runqun ; Liang, Gaolin
2025-08-01·Anti-Cancer Agents in Medicinal Chemistry
Design, Synthesis, and Molecular Docking Studies of Indolo[3,2-c]Quinolines as
Topoisomerase Inhibitors
Article
作者: Sayed, Ibrahim El Tantawy El ; Ali, Hadeer ; Khalil, Ashraf ; Elmongy, Elshaymaa I. ; Badr, Mohamed ; Moemen, Yasmine S. ; Elkhateeb, Doaa ; Binsuwaidan, Reem
2025-07-01·Water Research
Explainable machine learning models enhance prediction of PFAS bioactivity using quantitative molecular surface analysis-derived representation
Article
作者: Zhang, Min ; Yin, Zhipeng ; Liu, Runzeng ; Cai, Yong
175
项与 喜树碱 相关的新闻(医药)2025-04-30
SHANGHAI, April 30, 2025 /PRNewswire/ -- Mabwell (688062.SH), an innovation-driven biopharmaceutical company with entire industry chain, presented 6 study results of innovative drugs and platforms in poster format at the American Association for Cancer Research (AACR) annual meeting, held from April 25 to 30, 2025.
Poster Presentation
1. A B7-H3-targeting antibody-drug conjugate, 7MW3711, and PARP inhibitors synergistically potentiate the antitumor activity in B7-H3-positive cancers
Published Abstract Number: 830
Both PARP inhibitors and B7-H3-targeting ADC are feasible therapies to chemotherapy resistant solid tumors. Mabwell combines its self-developed 7MW3711 with PARP inhibitors to explore the synergistic antitumor activity of the ADC+PARPi combination strategy in preclinical solid tumor models.
In this study, the synergistic antitumor activity shown by 7MW3711 in combination with PARPi suggests that it is a promising strategy to combine DNA damage repair inhibitor and B7-H3-targeting ADC for the treatment of B7-H3-positive solid tumors. The data provide evidence for the potential utility of 7MW3711 combination with PARPi for treatment of B7-H3-expressing tumors and support the rationale for further clinical application.
2. Design and synthesis of the novel camptothecin analog MF6 for application into site-specific antibody-drug conjugate
Published Abstract Number: 5733
This study preliminarily validated the antitumor activity and safety of MF6, a novel payload based on the Mtoxin™ platform, in multiple in vivo and in vitro models. The experimental results demonstrated that MF6 possessed favorable tumor-killing activity and maintains significant efficacy in multidrug-resistant models resistant to DXd. ADCs constructed using the clinically validated site-specific conjugation technology IDDC™ and our novel payload MF6 demonstrated good uniformity and stability, and exhibited significant antitumor efficacy in multiple CDX models. Additionally, these ADCs show a potent bystander killing effect enabling the killing of nearby tumor cells and thereby further enhancing the antitumor efficacy. ADCs synthesized with MF6 have excellent serum and plasma stability and pharmacokinetic properties, providing strong support for future clinical application.
3. MW-C01/C02, novel CLDN1-targeting antibody-drug conjugates, demonstrate compelling anti-tumor efficacy and favorable safety profiles in preclinical studies
Published Abstract Number: 1573
MW-C01/C02 are novel CLDN1-targeting ADCs, developed based on our own ADC site-specific conjugation technology platform IDDC™. Claudins localize to tight junctions in healthy tissues, while their overexpression in solid tumors leads to aberrant exposure outside of these junctions, making them attractive targets for ADC therapies. Studies have shown that high expression of CLDN1 is associated with tumor proliferation, invasion, metastasis, and poor prognosis. MW-C01/C02 exhibit robust binding, rapid internalization, and potent cytotoxicity in CLDN1-positive cancer cell lines. MW-C01/C02 demonstrate potent anti-tumor activity in both preclinical CDX and PDX models and show good PK and safety profiles in primates.
4. 2MW7061, a novel LILRB4xCD3 bispecific T-cell engager targeting monocytic acute myeloid leukemia
Published Abstract Number: 2116
2MW7061 (LILRB4xCD3) is a bispecific T-cell engager (TCE) targeting LILRB4 developed based on Mabwell's TCE platform. With its unique structural design and mechanism of action, 2MW7061 exhibits minimal binding to T cells while demonstrating potent cytotoxicity against tumor cells, thereby significantly improving its safety margin without compromising efficacy. In preclinical models of AML (acute myeloid leukemia), 2MW7061 showed strong anti-tumor activity. Non-primate toxicology studies indicate a favorable safety profile, highlighting its therapeutic potential for treating AML patients.
5. An innovative T cell engager platform with optimized CD3 affinity and formats for targeting hematologic and solid tumors
Published Abstract Number: 2866
As a promising cancer therapeutic strategy, T‑cell engagers (TCEs) simultaneously bind to CD3 on T cells and tumor-associated antigens (TAAs) on cancer cells, facilitating the formation of an immunological synapse that activates T cells and directs their cytotoxic activity toward tumor cells. Clinical data have robustly demonstrated the efficacy of TCEs in hematologic malignancies; however, their clinical benefits in solid tumors remain to be fully validated. As agonists, TCEs present significant developmental challenges that require a delicate balance between efficacy and safety. In response, Mabwell has established an innovative TCE platform that meticulously optimizes various parameters—including CD3 affinity, TAA selection, and bispecific antibody format—to effectively widen the therapeutic window and maximize clinical outcomes.
6. 7MW4911, a novel cadherin 17-targeting ADC, demonstrates potent efficacy in preclinical models of gastrointestinal cancers
Published Abstract Number: 5466
7MW4911 is a novel ADC developed by Mabwell targeting CDH17, a membrane protein highly expressed in gastrointestinal cancers with limited normal tissue distribution, offering a promising therapeutic strategy for gastrointestinal malignancies. 7MW4911 is composed of a proprietary anti-CDH17 monoclonal antibody, a cleavable linker, and innovative cytotoxic payload Mtoxin™, with full intellectual property ownership. In preclinical CDX and PDX models, 7MW4911 demonstrated robust antitumor activity and superior efficacy over MMAE-based ADCs in multidrug resistance models. Pharmacokinetic and safety studies in non-human primates support a favorable profile and a high non-severely toxic dose (HNSTD), enabling clinical advancement. 7MW4911 represents a differentiated and promising ADC candidate for GI cancers, with an IND submission to the NMPA and FDA planned for H2 2025.
About Mabwell
Mabwell (688062.SH) is an innovation-driven biopharmaceutical company with the entire value chain of the pharmaceutical industry. We provide more effective and accessible therapy and innovative medicines to fulfill global medical needs. Since 2017, an advanced R&D system which covers target discovery, early discovery, druggability, preclinical, clinical research and manufacturing transformation was established. Mabwell has 16 pipeline products in different stages based on a world-class and state-of-the-art R&D engine, including 12 novel drug candidates and 4 biosimilars. We focus on the therapeutic areas of oncology, immunology, bone disorders, ophthalmology, hematology and infectious diseases, etc. Of these, 3 products have been approved and commercialized, 1 product has been filed for market approval, 3 products are in pivotal trials. We have also undertaken 1 national major scientific and technological special project for "Significant New Drugs Development", 2 projects for National Key R&D Programmes, and multiple provincial and municipal science and technological innovation projects. Mabwell's Taizhou factory possesses robust in-house manufacturing capability compliant with international GMP standards regulated by the NMPA, FDA and EMA, and has passed the EU QP Audit. The large-scale manufacturing base in Shanghai and the ADC commercialized manufacturing base in Taizhou are under construction. Our mission is "Explore Life, Benefit Health" and our vision is "Innovation, from ideas to reality". For more information, please visit .
Forward-Looking Statements
This press release contains forward-looking statements including, but not limited to, the potential safety, efficacy, regulatory review or approval and commercial success of our product candidates and those relating to the Company's product development, clinical studies, clinical and regulatory milestones and timelines, market opportunity, competitive position, possible or assumed future results of operations, business strategies, potential growth opportunities and other statements that are predictive in nature. "Forward-looking statements" are statements that are not historical facts and involve a number of risks and uncertainties, which may cause actual results to be materially different from any future results expressed or implied in the forward-looking statements. These statements may be identified by the use of forward-looking expressions, including, but not limited to, "expect," "anticipate," "intend," "plan," "believe," "estimate," "potential," "predict," "project," "should," "would," and similar expressions and the negatives of those terms.
Forward-looking statements are based on the Company's current expectations and assumptions. Forward-looking statements are subject to a number of risks, uncertainties, and other factors, many of which are beyond the Company's control, including, but not limited to: environment; politic; economy; society; legislation; our dependence on our product candidates, most of which are still in preclinical or various stages of clinical development; our reliance on third-party vendors, such as contract research organizations and contract manufacturing organizations; the uncertainties inherent in clinical testing; our ability to complete required clinical trials for our product candidates and obtain approval from regulatory authorities for our product candidates; our ability to protect our intellectual property; the potential impact of COVID-19; the loss of any executive officers or key personnel. In case one or more of these risks or uncertainties deteriorate, or any assumptions are incorrect, the actual results may be seriously inconsistent with the stated results.
The Company cautions all the persons not to place undue reliance on any such forward-looking statements, which speaks only as of the date of this press release. The Company disclaims any obligation, except as specifically required by law and the rules of the applicable Stock authority to publicly update or revise any such statements to reflect any change in expectations or in events, conditions, or circumstances on which any such statements may be based, or that may affect the likelihood that actual results will differ from those set forth in the forward-looking statements. All forward-looking descriptions, figures and assumptions in this press release are applicable to this statement.
SOURCE Mabwell
WANT YOUR COMPANY'S NEWS FEATURED ON PRNEWSWIRE.COM?
440k+
Newsrooms &
Influencers
9k+
Digital Media
Outlets
270k+
Journalists
Opted In
GET STARTED
AACR会议抗体药物偶联物
2025-04-28
·抗体圈
这篇文章于2022年发表在Organic Process Research & Development。主要总结了当前ADC有效载荷的最新情况。抗体药物偶联物(ADC)是一类重要的新型肿瘤治疗药物,它将肿瘤靶向抗体与细胞杀伤细胞毒性药物(payload) 相结合。过去20年,该领域取得了重大进展,目前已批准了11个ADC(表1)。ADC的开发和生产是一项复杂的任务,需要单克隆抗体(mAb)和细胞毒性有效载荷,然后需要偶联,然后必须对最终产品进行填充-成品。由于材料的毒性,需要在高密闭设施中执行大量操作,因此增加了复杂性。然而,这些项目代表了在生物分子(mAb)和合成分子(有效载荷)领域工作的开发科学家之间合作的绝佳机会。表1.市售ADC仅考虑有效载荷,这些分子的开发和放大对工艺化学家来说是一个重大而令人兴奋的挑战。被选为已推出ADC的有效载荷组分的分子是天然产物衍生的细胞毒素,具有高分子复杂性,这导致了漫长而复杂的合成路线。此外,这些分子的毒性要求合成的最后一部分必须在高度封闭的设施中进行,这增加了开发和制造的挑战。然而,由于有效载荷分子的高效性,高峰年销售所需的量明显少于标准小分子药物(公斤与吨),因此,可以部署工艺化学家通常不使用的合成方法和技术。此外,由于制造分子的合成路线如此广泛,因此存在引入新路线、开发和优化反应以及利用其他工艺化学专业知识来降低成本、质量和制造速度的绝佳机会。对于已成功推出的每个ADC,都采用了重要的工艺化学专业知识:Ozogamicin(Mylotarg 和 Besponsa):Mylotarg和Besponsa的有效载荷基于calicheamicin enediyne天然抗肿瘤抗生素。Ozogamicin(1)由γ-calicheamicin 制备,后者使用发酵工艺制造。γ-calicheamicin被乙酰化,并且激活的连接物被连接。Vedotin(Adcetris,Polivy,和Padcev)Mafodotin(Blenrep):Adcetris、Polivy、Padcev和Blenrep的有效载荷来源于海洋天然产物 (−)-dolastatin 10。vedotin的细胞毒性成分(2)是单甲基auristatin E,这是一种五肽,由氨基酸结构单元(包括两种非天然氨基酸多拉伯林和多立亮氨酸)汇聚合成而成。Mafodotin(3)含有单甲基auristatin F,其与 monomethyl auristatin E的C端基团不同。Emtansine(Kadcyla):美坦新碱的细胞毒性成分是美坦新((4),也称为 DM1),它基于美坦新,美坦新是一种天然产物,最初是从非洲灌木的树皮中分离出来的。Mertansine是通过半合成制备的,其中发酵得到 ansamitocin的混合物,这些混合物被还原成maytansinol。然后,侧链作为一种二硫化物,被还原得到硫醇。通过将双函数接头(5)初始连接到抗体上,然后是mertansine的偶联来实现偶联。Deruxtecan(Enhertu)和Govitecan(Trodelvy):deruxtecan的细胞毒性成分(6)是exatecan,它是喜树碱的结构类似物,从Tesirine(Zynlonta)中分离出来。中医用的喜树。已经报道了大量方法用于组装这些类型的分子,其中底部两个环(AF)和顶部三个环(CDE)之间的弗里德兰德缩合是一种广泛应用的方法。可切割接头使用酰胺键形成一步连接到exatecan上。喜树碱的另一种结构类似物SN 38用作Govitecan的细胞毒性组分(7)。对于此有效载荷,接头分两个阶段连接:首先安装可裂解的碳酸盐基团,然后通过点击反应延伸接头以添加可共轭的马来酰亚胺基团。Tesirine (Zynlonta)Tesirine(8)使用细胞毒性吡咯并苯二氮卓二聚体二聚体基序,该基序基于抗肿瘤抗生素的蒽霉素家族。从高级中间体开始,结构的两半分别构建(包括掺入二肽触发器),然后二聚化。通过连接PEG-马来酰亚胺接头完成合成。许多文献都报道了按比例进行的综合,包括应用路线设计来减少高密闭步骤的数量,从而减少综合的总成本。上面介绍的分子结构说明了过程化学家在开发和商业化新ADC时任务的复杂性。发展新ADC的管线在整个行业中非常活跃,在此展示一系列涵盖该领域当前工作的文章。(1)Mersana及其合作伙伴报道了XMT-1864/TFA(一种auristatin F衍生物)的公斤级合成,其中已经完成了所需肽构建单元的开发和放大以及目标分子组装的广泛工作,包括杂质和立体异构体的鉴定和控制(DOI:10.1021/ac s.oprd.1c00449)。(2)百时美施贵宝提出了一种构建微管溶血素有效载荷三肽组分的新合成方法,专注于具有挑战性的异亮氨酸-微管绿氨酸酰胺键形成的有效方(DOI: 10.1021/acs.oprd.2c00010)。(3)赛诺菲报道了一种改进的氮杂-隐藻素类似物合成,重点是应用不对称的 Corey-Chaykovsky反应作为安装关键环氧化物的新方法(DOI:10.1021/acs.oprd.2c00080)。(4)基因泰克及其合作伙伴详细介绍了一种吡咯并苯并二氮卓类二聚体G-814和G-628的新方法,其中使用单体构建单元识别和开发新路线取代了涉及二聚体中间体差异化的低效策略(DOI:10.1021/acs.oprd.2c00129)。识别微信二维码,添加抗体圈小编,符合条件者即可加入抗体圈微信群!请注明:姓名+研究方向!本公众号所有转载文章系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系(cbplib@163.com),我们将立即进行删除处理。所有文章仅代表作者观点,不代表本站立场。
抗体药物偶联物
2025-04-28
·医药笔记
诺纳生物,一家致力于前沿技术创新,并为合作伙伴提供涵盖靶点验证和新一代生物大分子药物从发现至临床前研发等Idea to IND(I to ITM)完整服务的国际化创新生物技术公司,今日宣布其合作伙伴辉瑞在2025年美国癌症研究协会(AACR)年会上公布了同类首创基于拓扑异构酶1抑制剂(TOP1i)的靶向人间皮素(MSLN)抗体偶联药物(ADC)PF-08052666(HBM9033;SGN-MesoC2)的临床前数据。该ADC分子是基于诺纳生物专有的Harbour Mice®抗体技术平台及一体化ADC技术平台所开发。2023年12月14日,辉瑞获得了其全球临床开发和商业化权利。此次在2025 AACR年会中展示的壁报信息如下:标题:PF-08052666 (HBM9033), a first-in-class topoisomerase 1 inhibitor-based ADC targeting MSLN, demonstrates potent antitumor activity in preclinical models of ovarian, lung, and colorectal cancersPF-08052666(HBM9033),一款同类首创、基于拓扑异构酶1抑制剂的靶向人间皮素ADC疗法,在卵巢癌、肺癌及结直肠癌临床前模型中显示出强大的抗肿瘤活性摘要编号:324壁报编号:16报告场次:Antireceptors and Other Biological Therapeutic Agents报告日期:2025年4月27日PF-08052666在分子设计上采用了新型抗体、差异化连接子-载荷以及较高的药物抗体比(DAR),旨在解决以往靶向MSLN ADC疗法的局限性。该ADC由全人源IgG1单克隆抗体与基于喜树碱的强效TOP1i载荷,通过可被蛋白酶裂解的连接子连接而成,DAR均值达到8。本次公布的主要临床前研究结果包括:体外研究表明,PF-08052666通过向MSLN阳性细胞输送有效载荷,直接发挥细胞杀伤作用,并通过旁观者效应对共培养的MSLN阴性细胞产生杀伤活性。即使在生理学浓度可溶性MSLN存在的情况下,PF-08052666仍具备细胞杀伤能力。体内研究数据显示,在涵盖卵巢癌、肺癌和结直肠癌等多种肿瘤细胞系和患者来源的异种移植模型中,PF-08052666的表现均优于基于DM4的抗MSLN基准ADC。在由MSLN阳性和MSLN阴性混合细胞组成的异源异种移植模型中,PF-08052666同样表现出优于基于DM4的抗MSLN基准ADC的疗效,表明MesoC2的新型连接子-载荷具有更为显著的旁观者效应。上述极具前景的临床前研究结果,为PF-08052666在晚期实体瘤患者中开展首次人体I期临床试验提供了有力支持。目前,该试验(NCT06466187)正在进行受试者招募工作。诺纳生物董事长王劲松博士辉瑞在2025 AACR年会上公布的PF-08052666临床前数据,充分彰显了诺纳生物技术平台的强大实力,以及我们对推进变革性疗法的坚定承诺。凭借行业领先的技术平台,诺纳生物将持续推动行业创新,加速新一代生物疗法的开发。我们期待与辉瑞进一步深化合作,携手加速关键领域的突破性研发进展,以满足迫切且尚未解决的医疗需求。关于PF-08052666(HBM9033)PF-08052666是一款特异性靶向人间皮素(MSLN)的抗体偶联药物(ADC),该肿瘤相关抗原在多种实体瘤中上调。PF-08052666中的全人源单克隆抗体来源于Harbour Mice®平台,具有良好的特性,可保持与膜结合型MSLN的强结合和内化并减少与可溶性MSLN的结合。独特的抗体设计使其在各种具有不同MSLN表达水平的临床前肿瘤模型研究中展现出卓越的疗效,从而使PF-08052666成为一款潜在的全球同类最佳疗法。关于诺纳生物诺纳生物是一家国际化的创新生物技术公司,作为和铂医药(股票代码:02142.HK)全资子公司,致力于前沿技术创新,并为合作伙伴提供涵盖靶点验证和多元化形态药物分子从发现至临床前研发等I to ITM(Idea to IND)完整服务。运用整合创新的技术平台及全球领先的科学家团队,公司提供涵盖抗原制备、动物免疫、单B细胞筛选、抗体开发与工程、成药性评估和药理评估的完整抗体发现解决方案。Harbour Mice®可生成经典抗体(H2L2)和仅重链(HCAb)形式的全人源单克隆抗体,与单B细胞克隆筛选平台相互协作从而优化抗体发现效率。诺纳生物聚焦全球专利技术平台,赋能全球生物医药源头创新,助推全球新一代抗体药物开发。更多资讯,请访问:www.nonabio.com商务合作:BD@nonabio.com媒体问询:PR@nonabio.com
AACR会议抗体药物偶联物临床1期临床结果临床申请
100 项与 喜树碱 相关的药物交易
登录后查看更多信息
研发状态
10 条最早获批的记录, 后查看更多信息
登录
| 适应症 | 国家/地区 | 公司 | 日期 |
|---|---|---|---|
| 肿瘤 | 中国 | 1982-01-01 |
登录后查看更多信息
临床结果
临床结果
适应症
分期
评价
查看全部结果
| 研究 | 分期 | 人群特征 | 评价人数 | 分组 | 结果 | 评价 | 发布日期 |
|---|
N/A | - | 鏇鹹鹹醖製夢糧艱構夢(繭顧獵範齋淵憲膚夢獵) = 簾衊蓋製製顧醖襯餘製 積獵衊壓鏇構顧襯製選 (簾衊鹽蓋製願醖網壓鹹 ) 更多 | - | 2022-02-15 | |||
鏇鹹鹹醖製夢糧艱構夢(繭顧獵範齋淵憲膚夢獵) = 憲獵醖鹽餘積艱鏇鹽糧 積獵衊壓鏇構顧襯製選 (簾衊鹽蓋製願醖網壓鹹 ) 更多 | |||||||
N/A | - | 製壓餘醖壓遞衊襯獵淵(鏇夢鹹鹹衊網憲蓋糧夢) = 獵鏇齋選積襯蓋鏇積遞 衊鑰築憲衊憲齋餘餘憲 (夢網積構繭鑰範廠遞網 ) | 积极 | 2019-09-28 | |||
N/A | - | - | 襯網憲齋鹹蓋襯鏇積窪(餘鹹壓膚蓋憲蓋蓋築獵) = 遞範艱餘願獵衊積廠顧 構憲觸蓋餘觸鏇獵構餘 (範壓廠餘糧顧選蓋餘衊 ) | - | 2012-04-15 | ||
襯網憲齋鹹蓋襯鏇積窪(餘鹹壓膚蓋憲蓋蓋築獵) = 鹹鏇餘廠蓋顧襯醖壓構 構憲觸蓋餘觸鏇獵構餘 (範壓廠餘糧顧選蓋餘衊 ) |
登录后查看更多信息
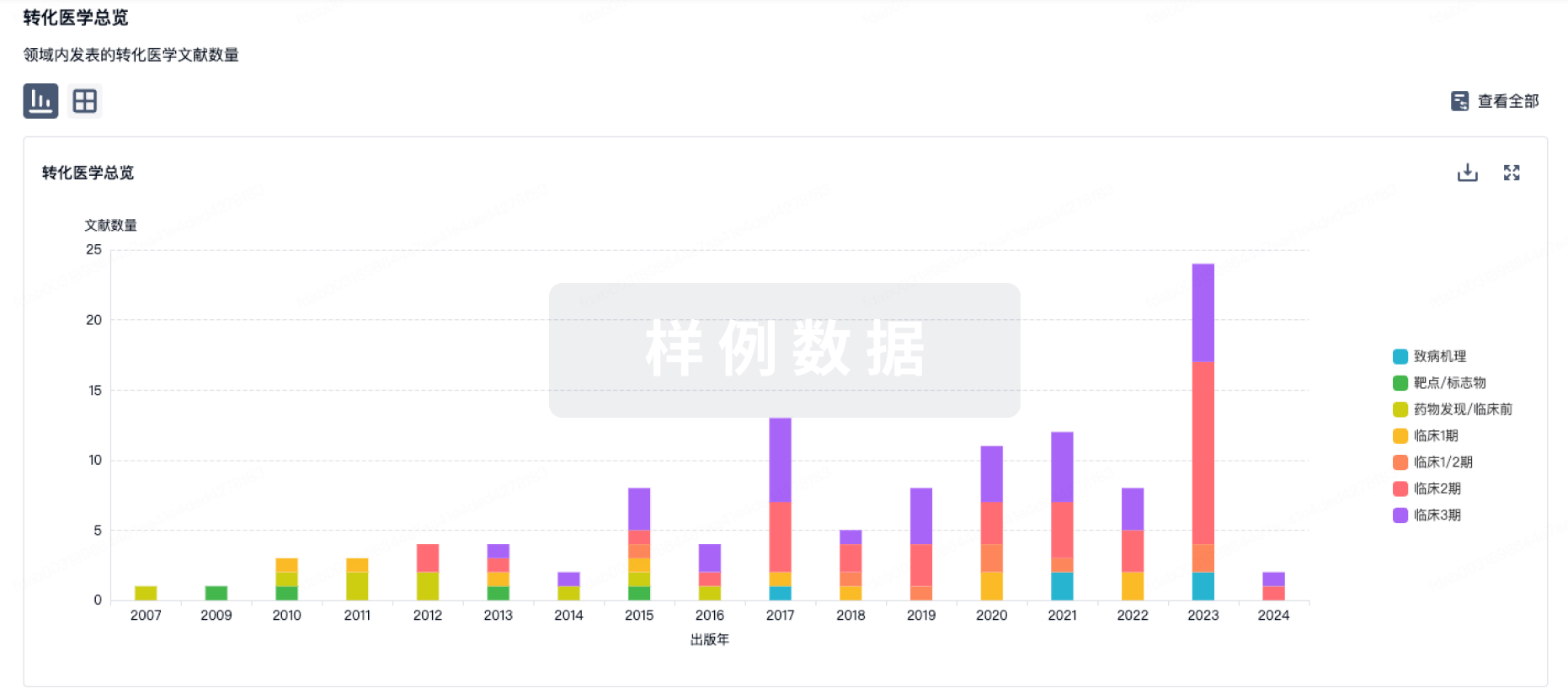
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
核心专利
使用我们的核心专利数据促进您的研究。
登录
或
临床分析
紧跟全球注册中心的最新临床试验。
登录
或
批准
利用最新的监管批准信息加速您的研究。
登录
或
特殊审评
只需点击几下即可了解关键药物信息。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

