预约演示
更新于:2025-05-07
KRAS mutation-related tumors
KRAS突变肿瘤
更新于:2025-05-07
基本信息
别名 CFC2 mutation-related tumors、K-RAS2A mutation-related tumors、K-RAS2B mutation-related tumors + [10] |
简介- |
关联
132
项与 KRAS突变肿瘤 相关的药物作用机制 ROS1抑制剂 [+3] |
在研适应症 |
非在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2023-11-15 |
作用机制 MEK1抑制剂 [+1] |
非在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2020-04-10 |
作用机制 MEK1抑制剂 [+1] |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2018-06-27 |
36
项与 KRAS突变肿瘤 相关的临床试验NCT06707896
Phase I, Open-Label Study of Autologous Mutant KRAS-redirected T-cell Receptor Cells (TCR1020-CD8)
This is a Phase I, open-label dose finding study to assess the safety, manufacturing feasibility, and preliminary efficacy of TCR1020-CD8 T cells in patients with KRAS-mutated cancers. Initially, patients with KRAS G12V mutation positive metastatic pancreatic adenocarcinoma or colorectal cancer will be targeted for participation. Up to 4 total dose levels will be evaluated using a 3+3 dose escalation design.
开始日期2025-06-01 |
申办/合作机构 |
NCT06917079
A Phase 1a/1b Open-Label Study Evaluating the Safety, Tolerability, Pharmacokinetics, and Efficacy of BBO-11818 in Subjects With Advanced KRAS Mutant Cancers
A first in human study to evaluate the safety and preliminary antitumor activity of BBO-11818, a pan-KRAS inhibitor, in subjects with locally advanced unresectable or metastatic KRAS mutant solid tumors.
开始日期2025-03-31 |
申办/合作机构 |
NCT06456138
Trametinib Plus Anlotinib Combined With Tislelizumab in KRAS-mutant Advanced Non-small Cell Lung Cancer Patients: a Multi-center, Open-label, Phase 1/2 Study
Lung cancer is the most common cause of cancer-related death worldwide. Approximately 85% to 90% of lung cancer cases are non-small cell lung cancer (NSCLC), of which KRAS is one of the most common driver genes, occurring in 25-30% of lung adenocarcinomas and 3-5% of squamous cell carcinomas. KRAS-mutant NSCLC had been considered undruggable in past decades. This research sought to address a significant challenge in treating NSCLC with KRAS mutations, which are notoriously difficult to target effectively. Here, we proposal that the combined use of anlotinib and trametinib combined with tislelizumab may form an effective strategy for the treatment of KRAS-mutant NSCLC patients.
开始日期2024-07-01 |
申办/合作机构  上海市胸科医院 上海市胸科医院 [+3] |
100 项与 KRAS突变肿瘤 相关的临床结果
登录后查看更多信息
100 项与 KRAS突变肿瘤 相关的转化医学
登录后查看更多信息
0 项与 KRAS突变肿瘤 相关的专利(医药)
登录后查看更多信息
556
项与 KRAS突变肿瘤 相关的文献(医药)2025-04-24·Journal of Medicinal Chemistry
Copper-KRAS-COX2 Axis: A Therapeutic Vulnerability in Pancreatic Cancer
Article
作者: Hanafi, Maha ; Neamati, Nouri ; Xu, Yibin ; Kyani, Armita ; Wen, Bo ; Mouawad, Rima ; Roy, Joyeeta ; Sun, Duxin
2025-04-02·Molecular Cancer Therapeutics
Pan-KRAS Inhibitors BI-2493 and BI-2865 Display Potent Antitumor Activity in Tumors with KRAS Wild-type Allele Amplification
Article
作者: Geyer, Antonia ; Rocchetti, Francesca ; Boghossian, Andrew S. ; Samwer, Matthias ; Ronan, Melissa M. ; Popow, Johannes ; Ebner, Florian ; Lipp, Jesse ; Gmachl, Michael ; Gerlach, Daniel ; Roth, Jennifer A. ; Rees, Matthew G. ; Tedeschi, Antonio ; Schischlik, Fiorella ; Rudolph, Dorothea ; Pearson, Mark ; Santoro, Valeria ; Kraut, Norbert
2025-04-01·ESMO Open
Pan-cancer clinical and molecular landscape of MTAP deletion in nationwide and international comprehensive genomic data
Article
作者: Oda, K ; Kage, H ; Shinozaki-Ushiku, A ; Ikushima, H ; Watanabe, K
309
项与 KRAS突变肿瘤 相关的新闻(医药)2025-05-01
·小药说药
-01-引言肿瘤是一个复杂的生态系统,其中癌症细胞和多种宿主细胞之间的相互作用影响疾病的进展和治疗反应。在肿瘤进展过程中,癌症细胞采用多种途径来逃避免疫攻击,例如下调抗原提呈机制或诱导抑制性免疫检查点分子,而免疫压力在肿瘤的发展和转移扩散过程中促进克隆进化。同时,癌症细胞劫持免疫细胞,如中性粒细胞、巨噬细胞和调节性T细胞(Treg),以协调免疫抑制性肿瘤微环境(TME)。这反过来促进免疫逃逸,促进血管和细胞外基质的重塑,并支持癌症进展和治疗抵抗。异常免疫反应被广泛认为是癌症的标志,并为癌症治疗提供了有吸引力的靶点。因此,我们有必要深入了解癌症细胞的内在特征,包括遗传变异、信号通路调节和代谢改变,它们在协调免疫景观的组成和功能状态方面发挥关键作用,并影响免疫调节策略的治疗效益。-02-一、遗传改变癌基因、抑癌基因或DNA损伤修复基因中的特定癌症细胞固有遗传改变有助于调节癌症免疫状况。癌基因癌症细胞中不同的致癌基因通过不同的机制调节免疫行为,这些机制因癌症类型、癌症分期和癌症部位而异。例如,KRAS突变通过IL-8诱导的炎症、IL-6介导的巨噬细胞和Treg细胞浸润、GM-CSF诱导的Gr1+CD11b+髓系细胞的扩增、IL-10和TGF-β介导的CD4+CD25−T细胞向Treg细胞的转化、以及抑制干扰素调节因子2(IRF2)和随后产生的CXCL3,从而导致CD8+T细胞抑制,髓系衍生抑制细胞(MDSCs)的迁移增强。MYC通过增强CD47和PD-L1表达来抑制抗肿瘤免疫,从而削弱巨噬细胞和T细胞的募集。此外通过分泌IL-1β,阻断细胞毒性T细胞、NK细胞和树突状细胞的浸润,并增强支持肿瘤的巨噬细胞和中性粒细胞的浸润。KRAS突变和MYC还通过与MYC相互作用的锌指蛋白1(MIZ1)产生协同作用,介导IFN-β的抑制以及CCL9和IL-23的分泌,增强PD-L1+巨噬细胞浸润,抑制B细胞、T细胞和NK淋巴细胞驱动免疫逃避。此外,突变的表皮生长因子受体(EGFR)通过降低PD-L1表达来驱动免疫逃逸,通过IRF1介导的CXCL10下调阻碍CD8+T细胞募集,并通过CD73依赖性腺苷生成或JNK–JUN介导的CCL2上调促进Treg细胞浸润。人表皮生长因子受体2(HER2)扩增导致主要组织相容性复合物I类(MHC-I)的下调和TANK结合激酶1(TBK1)的磷酸化,其抑制下游cGAS–STING信号传导和IFNβ产生,导致免疫逃避。抑癌基因抑癌基因通过不同的环境依赖机制调节免疫。例如,研究发现Trp53的缺失通过WNT配体介导的巨噬细胞极化和IL-1β的产生驱动免疫抑制,导致中性粒细胞的全身聚集,从而CD8+T细胞的抑制和CXCL17、CCL3、CCL11、CXCL5和M-CSF介导的巨噬细胞和Treg细胞的募集。TP53的突变与PD-L1表达相关,并通过CXCL2分泌调节免疫抑制中性粒细胞的募集,通过NF-κB介导的IL-8调节慢性炎症,并且结合TBK1抑制下游cGAS–STING–IRF3信号传导和IFN-β1的产生,其减少T细胞和NK细胞浸润并增强巨噬细胞极化。相反,TP53的功能增益突变与新抗原表达,以及IFN-γ和CXCL9表达相关,这支持抗肿瘤免疫。在KRAS突变肿瘤中,丝氨酸/苏氨酸激酶11(STK11)的失活通过粒细胞集落刺激因子(G-CSF)、IL-6和CXCL7的表达刺激免疫抑制中性粒细胞的积聚。此外,KRAS和STK11突变肿瘤中PD-L1的表达会导致对免疫检查点阻断(ICB)的抵抗。最后,PTEN丢失通过CCL2和VEGF的分泌减少CD8+T细胞浸润,并激活PI3K信号增强PD-L1的表达,以及NF-κB介导的CCL20、CXCL1、IL-6和IL-23分泌增强Treg细胞和髓系细胞浸润,并通过IL-1β和M-CSF驱动局部MDSC扩增。DNA损伤修复机制的遗传改变癌症细胞中的错配修复(MMR)缺陷会导致突变、新抗原和细胞中DNA的积累,从而触发cGAS–STING依赖性的T细胞浸润,免疫检查点分子如PD-1、PDL1、CTLA-4和LAG-3在浸润免疫细胞上的表达增强。在BRCA1突变的肿瘤中,双链DNA积聚在细胞中,并引发与CD8+T细胞浸润相关的cGAS–STING介导的炎症反应。cGAS–STING还促进免疫抑制性髓系细胞、巨噬细胞、Treg细胞和耗竭的PD1+T细胞的募集,以及VEGF的上调。DNA传感基因和下游介质(如IFN-β或CCL5)的遗传缺失或表观遗传沉默可介导BRCA1突变肿瘤的免疫逃逸,以及PD-L1的表达增强。相反,BRCA2突变肿瘤显示cGAS–STING介导的各种T细胞群的富集。-03- 二、表观遗传修饰除了遗传改变外,表观遗传改变也是癌症细胞的一个共同特征,在核结构和基因转录中起着至关重要的作用。癌症表观基因组会发生各种变化,包括DNA甲基化、组蛋白修饰、染色质重塑和非编码RNA的失调。越来越多的证据表明,癌症细胞中发生的表观遗传变化也会影响与免疫系统的串扰。DNA甲基化由于超级增强子(SE)的形成,炎症基因的DNA去甲基化导致CXC趋化因子配体(CXCL)介导的中性粒细胞募集,而总体低甲基化与PD-L1、IL-6和VEGF的表达增强相关。而DNA(超)甲基化可降低编码PD-L1的基因CD274和编码MHC-I的HLA基因的表达,而编码cGAS–STING的基因通过启动子甲基化的转录抑制增强MHC-I表达和T细胞识别。异柠檬酸脱氢酶1(IDH1)或IDH2的突变通过将α-酮戊二酸转化为(R)-2-羟基戊二酸(2HG)来诱导全局超甲基化。而编码PD-L1、CXCL9和CXCL10免疫相关基因的转录抑制会损害IDH突变肿瘤中CD8+T细胞的浸润,G-CSF的转录激活会增强非抑制性中性粒细胞和前中性粒细胞的浸润。组蛋白甲基化组蛋白H3在Lys27发生三甲基化(H3K27me3),在DNA低甲基化的背景下,抑制编码MHC-I、CXCL9和CXCL10的基因,并减少CD8+T细胞浸润。癌症细胞中的多梳抑制复合物2(PRC2)活性介导H3K27me3,抑制G-CSF和IL-6的表达并诱导性一氧化氮合成酶阳性(iNOS+)巨噬细胞、中性粒细胞和T细胞的招募。ARID1A是SWI/SNF复合物的DNA结合亚基, ARID1A的遗传突变会抑制IFN-γ诱导基因转录和CXCL9、CXCL10和CXCL11的表达,从而抑制T细胞浸润。组蛋白乙酰化组蛋白乙酰化通过组蛋白乙酰转移酶1(HAT1)调节免疫应答,HAT1介导CD274和组蛋白去乙酰化酶(HDACs)的表达,HDACs抑制编码PD-L1和PD-L2的基因的表达,以及CCL5、CXCL9和CXCL10的表达,从而损害T细胞浸润。-04-三、细胞内信号通路癌症细胞的一个关键特征是异常信号传导,这是由编码或非编码DNA区域的遗传或表观遗传改变以及生长因子和代谢等外部信号驱动的。WNT–β-catenin通路激活的癌症细胞内固有WNT–β-catenin信号与免疫排斥相关,但其潜在机制仍不明确。WNT–β-catenin通过诱导ATF3表达阻断CD103+树突状细胞的募集和T细胞启动,并抑制CCL4或CCL5分泌。TGF-β通路淋巴细胞抗原LY6K和LY6E在癌症细胞中的过表达激活TGF-β信号转导和SMAD2/3磷酸化,导致NK细胞活性降低和Treg细胞浸润增强。αVβ6整合素的上调通过激活TGF-β和SMAD2/3磷酸化来调节SOX4的表达,SOX4抑制多种I型干扰素诱导基因和编码MHC-I的基因,同时增强PD-L1表达,这会损害细胞毒性T细胞介导的免疫。此外,癌症细胞衍生的TGF-β介导CD4+T细胞向Treg细胞的转化。而SMAD4的敲除会导致肿瘤细胞中TGF-β信号的失活,这增强了CCL9的表达和未成熟髓系细胞的募集。NF-κB信号通路PD-L1的表达受癌症细胞内NF-κB信号转导的转录调控,其途径是通过MUC1-c与CD274启动子上的NF-κB亚基p65的直接结合,或通过TGF-β介导的MRTFA的表达。MUC1-C和p65还驱动ZEB1转录,这导致编码CCL2、IFN-γ、GM-CSF和TLR9的基因受到抑制,CD8+T细胞浸润受损。相反,免疫疗法和化疗诱导的NF-κB和p300–CBP的活化增强了MHC-I抗原呈递和CD8+T细胞识别。HIF信号通路缺氧诱导的缺氧诱导因子1α(HIF1α)信号通路通过增强CD274的表达和抑制编码CCL2和CCL5的基因来驱动免疫逃避,这会损害NK细胞和CD8+T细胞的浸润,而HIF2α诱导CXCL1的表达并促进肿瘤的中性粒细胞募集。-05-四、代谢改变在肿瘤的发生和进展过程中,癌症细胞及其TME不断暴露在恶劣的条件和压力下。为了生存和维持生长,需要细胞适应和代谢重编程。癌症代谢重编程与免疫抑制和逃避有关。肿瘤细胞消耗大量葡萄糖,这与T细胞浸润不良有关。高葡萄糖消耗导致乳酸的产生并分泌到肿瘤微环境中,乳酸在肿瘤微环境中以免疫抑制的方式起作用,降低NK细胞的细胞溶解活性,并增强PD-1表达和Treg细胞的免疫抑制能力。此外,乳酸增加了肿瘤和脾脏中MDSC的频率,并在肿瘤相关巨噬细胞中诱导“M2样”极化。此外,癌症细胞中谷氨酰胺的消耗降低CD8+T细胞的活化和浸润,并通过增加G-CSF的分泌增强MDSC的募集。-06-结语近年来,关于癌症细胞-免疫细胞串扰的研究大幅增长。跨越式发展的技术进步,包括单细胞多组学技术,基于人工智能的系统生物学方法以及体内体细胞基因编辑策略,有望加速这些研究的深入,并最终形成针对个体患者的新型免疫干预策略。展望未来,个性化免疫干预策略将需要综合多组学肿瘤表征、免疫分析、液体活检样本的动态监测以评估基于血液的生物标志物、计算数据分析和转化,以及临床相关体外和体内模型的机制研究。有关癌症细胞内在和癌症细胞外部特征的知识体系将共同决定局部和系统免疫状况,以指导临床决策,并为针对患者个性化新型治疗方法开辟道路。参考资料:1.Mechanisms driving the immunoregulatory function of cancer cells. Nat Rev Cancer.2023 Jan 30.公众号内回复“ADC”或扫描下方图片中的二维码免费下载《抗体偶联药物:从基础到临床》的PDF格式电子书!公众号已建立“小药说药专业交流群”微信行业交流群以及读者交流群,扫描下方小编二维码加入,入行业群请主动告知姓名、工作单位和职务。
免疫疗法AACR会议临床1期
2025-04-28
- Preclinical data demonstrates synergy of QTX3544 with EGFR inhibitors, broadly enhancing anti-tumor activity - - Data supports ongoing Phase 1 clinical trial for QTX3544 as monotherapy and in combination with cetuximab in patients with KRASG12V mutations -
SOUTH SAN FRANCISCO, Calif., April 28, 2025 (GLOBE NEWSWIRE) -- Quanta Therapeutics, a privately-held clinical-stage biopharmaceutical company leading the development of innovative, oral therapeutics for RAS-driven cancers, today announced late-breaking preclinical data for QTX3544, an oral, G12V-preferring, dual ON/OFF state, multi-KRAS inhibitor currently being evaluated in a Phase 1 clinical trial in patients with KRASG12V mutation. Data are being presented at the American Association for Cancer Research (AACR) Annual Meeting 2025 being held April 25-30, 2025, in Chicago, Illinois. “There is a high unmet need for effective treatments in KRASG12V-mutated cancers, the second most prevalent in KRAS-driven cancers, for which there are no current targeted treatments, including colorectal and pancreatic cancers. Using a clinically validated approach against G12C mutations, we aim to combine allosteric KRAS inhibition and EGFR blockade to enhance the depth and durability of tumor response against G12V,” said Leonardo Faoro, MD, MBA, Chief Medical Officer of Quanta Therapeutics. “QTX3544 has shown high potency against multiple KRAS variants, particularly G12V. These preclinical data support the clinical strategy to combine QTX3544 and cetuximab to enhance the therapeutic activity against KRASG12V-driven cancers. We look forward to evaluating this combination in the ongoing Phase 1 clinical trial.” “We continue to generate promising data supporting our unique pipeline of KRAS-directed agents, as we advance three distinct candidates in the clinic targeting high prevalence KRAS mutations across solid tumors with major unmet need. QTX3034 and QTX3046 are being developed for G12D-mutant and QTX3544 for G12V-mutant KRAS-driven cancers,” said Perry Nisen, MD, PhD, Chief Executive Officer of Quanta. “We are building a compelling clinical package across our three lead programs and are on track to share proof-of-concept data later this year as our pipeline continues to progress.” Studies evaluating the potential synergy between QTX3544 and EGFR inhibitors (afatinib and cetuximab) in KRASG12V-driven cancer cell lines and tumor models demonstrated: QTX3544 synergized with EGFR inhibitors and potently inhibited KRASG12V-mutant cell proliferation in cell viability assays.Combined treatment of QTX3544 with afatinib led to a deeper suppression of MAPK signaling and enhanced inhibition of KRASG12V-driven cancer cell survival in colony formation assays.Combination treatment of QTX3544 with cetuximab significantly enhanced anti-tumor efficacy and elicited greater tumor regressions in pancreatic and colorectal KRASG12V-mutant cell line-derived xenograft models, as well as in a patient-derived colorectal tumor xenograft model. AACR presentation information:Title: QTX3544, a potent and selective G12V-preferring KRAS inhibitor, synergizes with EGFR inhibitors for enhanced anti-tumor activityDate and Time: Wednesday, April 30, 2025; 9:00 AM - 12:00 PMSession Name: Late-Breaking Research: Experimental and Molecular Therapeutics 4Abstract Number: LB430 Location: Poster Section 51 About QTX3544QTX3544 is an allosteric, G12V-preferring multi-KRAS inhibitor. The QTX3544 Phase 1 clinical trial is initially enrolling patients with KRASG12V mutation in dose escalation cohorts as monotherapy and in combination with cetuximab. The Phase 1 clinical endpoints include safety and tolerability, determination of the maximum tolerated dose/recommended Phase 2 dose, pharmacokinetic properties, anti-tumor activity, and molecular markers. The clinical trial is being conducted at clinical sites in the US. More information about the QTX3544 clinical trial (NCT06715124) can be found on https://clinicaltrials.gov/. About RAS and the MAPK PathwayThe mitogen-activated protein kinase (MAPK) pathway is a central signaling cascade that regulates cellular growth, proliferation, differentiation, and survival. When one of the proteins in the pathway is mutationally activated, it can drive tumor development and growth. RAS is the most frequently mutated oncogene in cancer, with KRAS mutations occurring in nearly one-quarter of all human cancers. RAS mutations impair the ability of RAS to convert from its active GTP-bound “ON” form into its inactive GDP-bound “OFF” state, leading to the sustained activation of the MAPK signaling pathway and ultimately driving tumorigenesis. KRAS mutations, especially G12D, G12V, and G12C, are highly prevalent in pancreatic, colorectal, and lung cancers. First-generation KRAS inhibitors have demonstrated clinical benefit, but their impact is limited to a subset of patients with a single type of KRAS mutation (G12C). About Quanta TherapeuticsQuanta Therapeutics is a private biopharmaceutical company focused on the most prevalent and elusive target in oncology—RAS. Our vision is to develop novel small molecule cancer medicines by selectively targeting protein-protein interactions that are key to oncogenic RAS activity. Driving Quanta's success is our unique high-throughput platform that applies Second Harmonic Generation (SHG) optical technology to identify allosteric modulators of protein complexes. The Quanta team has extensive drug development expertise and substantial research experience in the RAS space. By applying innovative medicinal chemistry and its unique protein conformation detection technology, Quanta aims to advance differentiated, next-generation RAS programs that address the resistance paradigms of targeted therapy in oncology. Quanta’s KRAS inhibitor pipeline includes three programs: QTX3034, a multi-KRAS inhibitor with G12D-preferring activity (G12D+ multi-KRAS), currently in a Phase 1 clinical trial as monotherapy and in combination with cetuximab; QTX3046, a G12D-selective KRAS inhibitor, currently in a Phase 1 clinical trial as monotherapy and in combination with cetuximab; QTX3544, a multi-KRAS inhibitor with G12V-preferring activity (G12V+ multi-KRAS) currently in a Phase 1 clinical trial as a monotherapy and in combination in patients with KRASG12V-driven solid tumors. Quanta is headquartered in South San Francisco, CA, and has a site in Radnor, PA. Find more information at https://www.quantatx.com/. Follow us on LinkedIn: Quanta Therapeutics. Quanta Therapeutics
Heather Meeks
661-992-6907
heather.meeks@quantatx.com
Media Contact
Kelli Perkins
kelli@redhousecomms.com
临床1期AACR会议
2025-04-28
劲方医药宣布公司自主开发的口服Pan RAS(ON)抑制剂GFH276临床前研究数据,于当地时间4月27日登陆美国癌症研究协会(AACR)年会壁报展示。GFH276为临床申报阶段的分子胶泛RAS抑制剂,可抑制多数野生/突变型RAS蛋白,在多种携带RAS突变或RTK改变的细胞及肿瘤模型中显示抑瘤活性,临床前研究还展现了GFH276对RAS下游MAPK信号通路的深度抑制。在多类KRAS突变肿瘤模型中,GFH276显示了相较于海外同类产品的更低起效剂量和更佳药代动力学特征。此外,GFH276在激酶选择性和安全相关靶点测试中展现良好的安全性及靶向特异性;并在多种机理诱导的KRAS抑制剂耐药细胞系中,均保持强效活性、显示多重抗耐药潜力。劲方研发团队向现场观众讲解(点击可见大图)摘要标题 | GFH276:Pan RAS (ON) 抑制剂临床前研究展现广谱抗肿瘤活性(摘要编号:389)抑制多数活化状态的野生/突变型RAS蛋白,并显示对RAS下游MAPK通路的深度抑制GFH276采用三元复合物(CypA-GFH276-RAS)作用机制,可抑制多数常见、活化状态的野生/突变型RAS蛋白,包括肿瘤中常见的KRAS突变型(G12C、G12D、G12V等)及NRAS、HRAS蛋白。此外,在多种KRAS突变或RTK(RAS通路上游蛋白)改变的细胞系中,GFH276均显示出有效的细胞生长抑制。RAS通过激活MAPK等下游信号通路,参与调控细胞增殖、迁移及存活等重要生理过程。在KRAS突变细胞及肿瘤模型中,GFH276均显示对MAPK通路ERK1/2蛋白磷酸化水平的迅速、深度抑制。相较于海外同类产品的更低起效剂量和更佳药代动力学特征在携带多种KRAS突变(G12C、G12D、G12V、G13D等)的非小细胞肺癌、胰腺癌、结直肠癌动物模型中,2-3周连续口服GFH276给药、剂量为0.3-3 mg/kg QD 即展现其剂量依赖式抑瘤活性;相较于同周期RMC-6236口服给药10 mg/kg QD的实验动物,GFH276口服给药1或3 mg/kg QD的实验动物即可实现同等或更大的肿瘤消退。GFH276的起效剂量优势与其优越的药代动力学特征相关:GFH276的在动物体内的清除率显著低于RMC-6236,口服生物利用度则显著高于后者。此外,GFH276在激酶选择性和安全相关靶点测试中未显示对非目标靶点的活性,展现其良好的安全性及靶向特异性。相较于SIIP结合式KRAS抑制剂的多重抗耐药潜力RTK重新激活可导致对KRAS抑制剂的适应性耐药出现;相较于第一代已上市KRAS抑制剂(SIIP结合式KRAS抑制剂),细胞实验显示EGF介导的RTK重激活几乎不影响GFH276对胞内ERK1/2磷酸化的抑制活性,显示了GFH276对抗适应性耐药的机制优势。此外,在多种SIIP(switch II pocket)结合式KRAS抑制剂的获得性耐药细胞模型中,GFH276仍显示有效的抗肿瘤细胞活性,展现其克服SIIP结合式KRAS抑制剂耐药局限的治疗潜力。 关于RAS蛋白及GFH276RAS蛋白为二元分子开关,在与GDP(二磷酸鸟苷)结合的失活状态和与GTP(三磷酸鸟苷)结合的活化状态之间切换,以此调控RAF-MEK-ERK、PI3K-AKT-mTOR等通路。RAS的致癌突变导致其 GTP 水解酶活性被破坏,从而令RAS蛋白主要以活化的GTP结合形式存在,会导致细胞恶性增殖和生物行为学的改变。RAS家族蛋白主要分为KRAS、HRAS、NRAS三大类,其中KRAS突变是肿瘤中最常见的基因突变之一。GFH276为机制独特的口服小分子Pan RAS(ON)抑制剂,采用三复合物作用机制(CypA-GFH276-RAS),可更高效抑制多数活化状态的野生/突变型RAS蛋白亚型,包括常见的KRAS突变型(G12C、G12D、G12V等)以及NRAS、HRAS亚型蛋白。临床前研究显示,GFH276呈现剂量依赖式的抗肿瘤活性并促进肿瘤消退;与第一代SIIP(switch II pocket)结合型KRAS抑制剂相比,GFH276有望克服现有药物导致的适应性、获得性耐药局限。 关于劲方医药 劲方医药是一家全球布局的创新药物开发企业,聚焦肿瘤、免疫类疾病领域高度未满足的临床需求,以疾病生物学机理和临床转化医学为核心,构建并发挥自主化、一体化研发体系优势,主攻尚无临床验证的创新靶点与适应症,并拥有全球自主知识产权。自2017年成立以来,劲方医药已建立包含多个自主研发的“全球新”大、小分子项目,多个产品在中国、欧洲、美国进入全球多中心临床试验,包括多项后期或关键性临床研究。达伯特®(氟泽雷塞)为劲方管线中首个国内上市获批产品,也是国内首个、全球第三个上市的KRAS G12C抑制剂,曾获得国家药品监督管理局优先审评及两项突破性疗法资格认定;氟泽雷塞、西妥昔单抗的一线联合疗法也在欧洲进入II期研究,为全球首个KRAS、EGFR抑制剂一线非小细胞肺癌联用治疗方案。目前,公司已形成RAS靶向疗法的一体化深耕矩阵,并积极拓展其他“全球新”靶向药及新类型疗法管线开发。同时,公司近年来不断深化商业合作网络,已与多个境内外上市公司达成战略授权协议、或开展“全球新”临床合作并已取得积极进展。 联系我们电话:021-68821388邮箱:pr@genfleet.com; bd@genfleet.com官网:www.genfleet.com 地址:上海市浦东新区张江路1206号A座邮编:201203
AACR会议临床结果
分析
对领域进行一次全面的分析。
登录
或
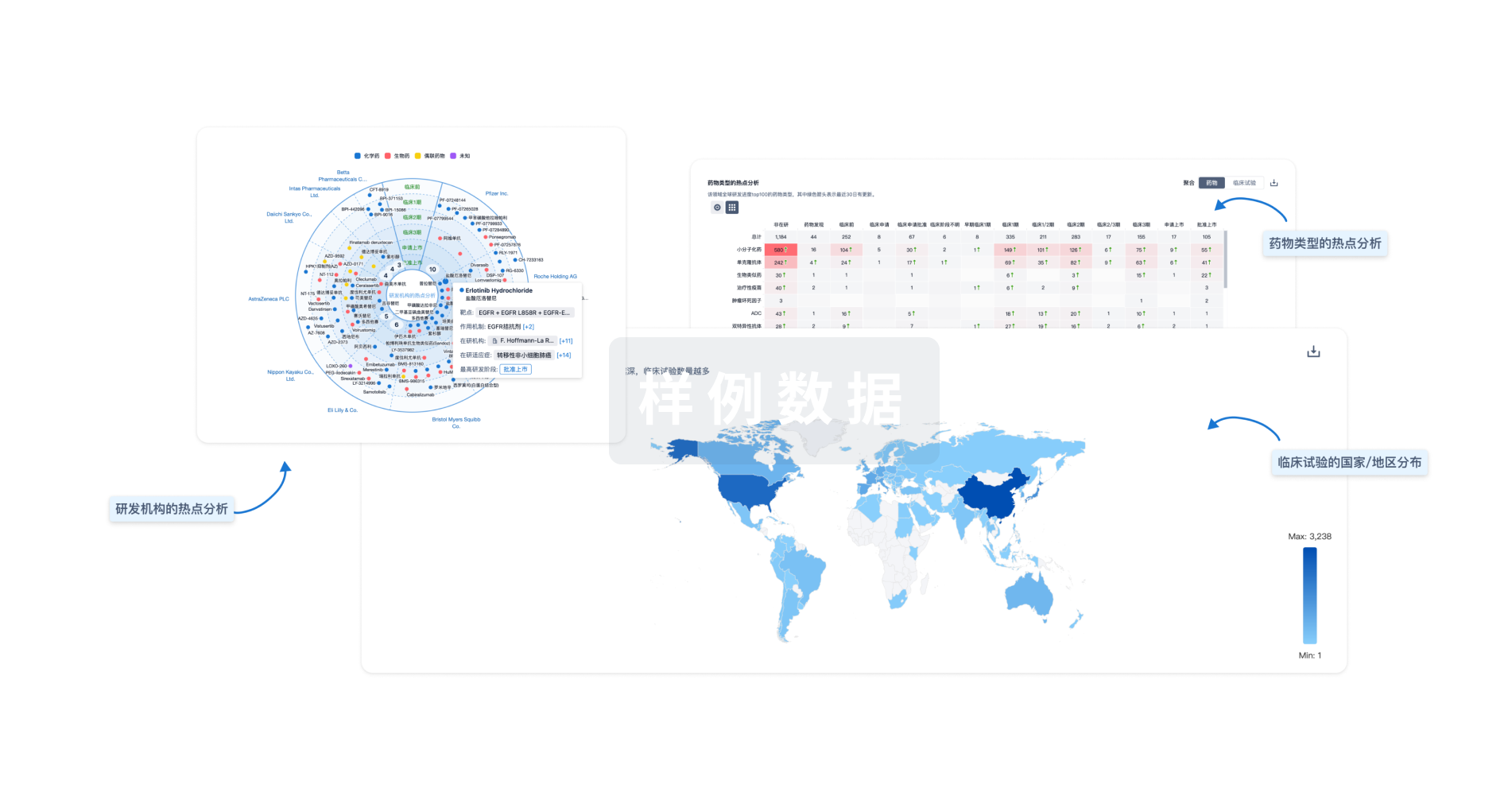
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用



