预约演示
更新于:2025-01-23
GRB10
更新于:2025-01-23
基本信息
别名 GRB10、GRB10 adapter protein、GRBIR + [4] |
简介 Adapter protein which modulates coupling of a number of cell surface receptor kinases with specific signaling pathways. Binds to, and suppress signals from, activated receptors tyrosine kinases, including the insulin (INSR) and insulin-like growth factor (IGF1R) receptors. The inhibitory effect can be achieved by 2 mechanisms: interference with the signaling pathway and increased receptor degradation. Delays and reduces AKT1 phosphorylation in response to insulin stimulation. Blocks association between INSR and IRS1 and IRS2 and prevents insulin-stimulated IRS1 and IRS2 tyrosine phosphorylation. Recruits NEDD4 to IGF1R, leading to IGF1R ubiquitination, increased internalization and degradation by both the proteasomal and lysosomal pathways. May play a role in mediating insulin-stimulated ubiquitination of INSR, leading to proteasomal degradation. Negatively regulates Wnt signaling by interacting with LRP6 intracellular portion and interfering with the binding of AXIN1 to LRP6. Positive regulator of the KDR/VEGFR-2 signaling pathway. May inhibit NEDD4-mediated degradation of KDR/VEGFR-2. |
关联
1
项与 GRB10 相关的药物靶点 |
作用机制 GRB10 antagonists |
在研机构- |
在研适应症- |
非在研适应症 |
最高研发阶段无进展 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
100 项与 GRB10 相关的临床结果
登录后查看更多信息
100 项与 GRB10 相关的转化医学
登录后查看更多信息
0 项与 GRB10 相关的专利(医药)
登录后查看更多信息
431
项与 GRB10 相关的文献(医药)2024-10-01·Cancer Epidemiology
Genetic variation near GRB10 associated with bone growth and osteosarcoma risk in canine and human populations
Article
作者: Wimberly, Courtney E ; Hansen, Helen M ; Lucas, Sydney E ; Morimoto, Libby M ; Ostrom, Quinn T ; de Smith, Adam J ; Wiemels, Joseph L ; Wagner, Lars M ; Parmar, Kajal V ; Yang, Tianzhong ; Graves, Laurie A ; Eward, William C ; Walsh, Kyle M ; Metayer, Catherine ; Spector, Logan G
2024-06-01·Poultry Science
Melatonin alleviates endoplasmic reticulum stress to improve ovarian function by regulating the mTOR pathway in aged laying hens
Article
作者: Su, Bo-fei ; Xue, Han ; Chang, Li-Yun ; Chen, Yi-fan ; Chang, Li-yun ; Liu, Xue-Lu ; Hao, Er-ying ; Wang, De-He ; Wang, De-he ; Shi, Lei ; Su, Bo-Fei ; Hao, Er-Ying ; Chen, Yi-Fan ; Liu, Xue-lu ; Chen, Hui
2024-06-01·Environmental Health Perspectives
Effects of Developmental Lead and Phthalate Exposures on DNA Methylation in Adult Mouse Blood, Brain, and Liver: A Focus on Genomic Imprinting by Tissue and Sex
Article
作者: Cavalcante, Raymond ; Perera, Bambarendage P.U. ; Colacino, Justin A. ; Wang, Kai ; Morgan, Rachel K. ; Dolinoy, Dana C. ; Jones, Tamara R. ; Neier, Kari ; Bartolomei, Marisa S. ; Sartor, Maureen A. ; Rygiel, Christine A. ; Lalancette, Claudia ; Prasasya, Rexxi ; Svoboda, Laurie K.
3
项与 GRB10 相关的新闻(医药)2023-01-30
·生物谷
基于印迹基因的QCIGISH检测在区分甲状腺结节良恶性上具有很高的诊断效能,其对恶性结节很高的阴性预测值证明其在排除恶性肿瘤上非常有效,同时,很高的阳性预测值可使得其作为一种有效的检测方法
甲状腺结节的患病率很高,在成人群体中以超声检查方式统计的患病率为70%,其中约5%的结节是恶性的[1],而良恶性结节的治疗方式及预后差别巨大。因此,准确判断甲状腺结节的良恶性对于其临床管理至关重要。
目前,超声影像学联合细针穿刺(FNA)活检是甲状腺结节诊断的主要方法,但约20%-30%的甲状腺结节仍无法通过这种方法确定性质[2]。因此,急需开发新的检测方法以提高其诊断精确度。
基于生物标志物的肿瘤检测,极大地改变了肿瘤检测的格局,其中基因组印迹作为肿瘤发生过程中的表观遗传标志物,与多种肿瘤的发生关系密切[3],然而这种基因印迹的检测却存在较高的技术壁垒。
近日,由南方科技大学邢明照教授领衔的研究团队在Journal of Clinical Oncology发表了一项重要研究成果。研究发现,基于印记生物标志物的检测可有效区分良恶性甲状腺结节,尤其对于甲状腺恶性结节,敏感性高达100%[4],表明基于印迹基因的新检测方法,在甲状腺结节的临床诊断方面具有很高的潜力。
图1. 文章封面截图
研究人员纳入了中国科学院大学肿瘤医院等8个大型医疗中心550名甲状腺结节患者的病理标本,其中124例(82例恶性结节和42例良性结节)为术后石蜡标本,用于初步诊断模型的构建,32例(8例恶性结节和24例良性结节)为术前甲状腺穿刺标本、用于模型优化,394(117例恶性结节和277例良性结节)例为术后组织病理学的穿刺样本,以进行前瞻性验证研究(图2)。
图2. 研究设计和流程图
由于印迹基因检测存在较高的技术难度,因此,如何完成这550例样本的印迹基因检测就成了研究的核心。
RNA原位杂交(ISH)方法是检测印迹和非印迹基因的转录调控的传统技术[5],为了精确定量印迹基因,研究人员基于这种传统技术,开发出一种更为先进的定量显色印迹基因原位杂交技术(QCIGISH),并鉴定出三个具有癌症诊断潜力的印迹基因:鸟嘌呤核苷酸结合蛋白-α刺激复合物位点(GNAS),生长因子受体结合蛋白(GRB10)和小核核糖核蛋白多肽N(SNRPN)[6]。
那基于QCIGISH检测这些癌症特异性印迹基因,是否可以准确判断出甲状腺结节的良恶性呢?
基于QCIGISH检测技术,研究人员使用靶向GNAS、GRB10、SNRPN和HM13非编码内含子序列的探针进行印迹基因检测,并测量其双等位基因表达(BAE)、多等位基因表达(MAE)和总表达(TE)来对印迹基因的灵敏性和特异性进行客观定量(图3A)。随后,研究人员通过结节组织连续切片进行QCIGISH检测和HE染色,以分析这四种印迹基因表达与甲状腺结节良恶性的相关性(图3B)。
图3. QCIGISH的原理-印迹基因等位基因表达状态的可视化、定量和病理确认
完成初始化模型构建和模型优化后,研究人员对394例用于前瞻性验证的临床样本进行了QCIGISH临床适用性的测试,发现超声检查ACR TI-RADS分类中3-5级结节中组织病理恶性肿瘤率随级别增加逐渐升高,Bethesda分型也发现级别越高恶性肿瘤率也越高。
随后,研究人员以组织病理学诊断作为标准,评估了QCIGISH分级模型的诊断效能:QCIGISH诊断甲状腺恶性结节的总体敏感性为100%,良性结节的特异性为91.5%,总体阳性预测值(PPV)为96.5%,恶性肿瘤的阴性预测值(NPV)为100%(图4),相应的总体诊断准确率为97.5%(384/394)。
图4. QCIGISH诊断甲状腺结节的诊断效能
接下来,研究人员评估了QCIGISH在不同Bethesda分型患者中的诊断效能:
对于Bethesda II型甲状腺结节,100%(83/83)的QCIGISH阴性病例为组织病理学良性,而42.9%(6/14)的QCIGISH阳性病例为组织病理学恶性(图5D);
对于Bethesda III和IV型甲状腺结节,QCIGISH阴性病例中100%(23/23)为组织病理学良性结节,QCIGISH阳性病例中96.6%(56/58)为组织病理学恶性结节,PPV为96.6%,NPV为100%(图5E),诊断恶性甲状腺结节的总体准确率为97.5%(79/81);
对于Bethesda V型甲状腺结节,仅一例为QCIGISH阴性且为组织病理学良性结节,而100%(31/31)的QCIGISH阳性病例为组织病理学恶性结节(图5F);
对于Bethesda VI型甲状腺结节,100%(184/184)的QCIGISH阳性病例为组织病理学恶性结节(图5G)。
图5. QCIGISH诊断不同Bethesda分型甲状腺结节的诊断效能
综上所述,基于印迹基因的QCIGISH检测在区分甲状腺结节良恶性上具有很高的诊断效能,其对恶性结节很高的阴性预测值证明其在排除恶性肿瘤上非常有效,同时,很高的阳性预测值可使得其作为一种有效的检测方法,以辨别细胞学上不确定的甲状腺结节。这项研究提供了一种新的甲状腺分子诊断检测方法,或可改变甲状腺临床诊断的格局。
参考文献:
[1] Guth S, Theune U, Aberle J, et al. Very high prevalence of thyroid nodules detected by high frequency (13 MHz) ultrasound examination. Eur J Clin Invest. 2009 Aug;39(8):699-706. doi: 10.1111/j.1365-2362.2009.02162.x.
[2] Wang CC, Friedman L, Kennedy GC, et al. A large multicenter correlation study of thyroid nodule cytopathology and histopathology. Thyroid. 2011 Mar;21(3):243-51. doi: 10.1089/thy.2010.0243.
[3] Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007 Feb;211(3):261-8. doi: 10.1002/path.2116.
[4] Xu H, Zhang Y, Wu H, et al. High Diagnostic Accuracy of Epigenetic Imprinting Biomarkers in Thyroid Nodules. J Clin Oncol. 2022 Nov 15:JCO2200232. doi: 10.1200/JCO.22.00232.
[5] Kohda A, Taguchi H, Okumura K. Visualization of biallelic expression of the imprinted SNRPN gene induced by inhibitors of DNA methylation and histone deacetylation. Biosci Biotechnol Biochem. 2001 May;65(5):1236-9. doi: 10.1271/bbb.65.
[6] Shen R, Cheng T, Xu C, et al Novel visualized quantitative epigenetic imprinted gene biomarkers diagnose the malignancy of ten cancer types. Clin Epigenetics. 2020 May 24;12(1):71. doi: 10.1186/s13148-020-00861-1.
2023-01-28
·生物谷
来自中国中南大学等机构的科学家们通过研究揭示了激素瘦素活性被调节的分子机制,瘦素是一种能参与控制机体食欲和能量消耗的特殊激素。
对很多人而言,保持健康的体重或许是一项挑战,据美国CDC数据显示,在美国20岁及以上的成年人群中,肥胖的流行率为42%,包括心脏病、中风、2型糖尿病和某些类型癌症在内的肥胖相关疾病是可预防的人群过早死亡的主要原因之一。为了寻找策略来帮助人们达到并维持健康体重,近日,一篇发表在国际杂志Nature Metabolism上题为“Hypothalamic Grb10 enhances leptin signalling and promotes weight loss”的研究报告中,来自中国中南大学等机构的科学家们通过研究揭示了激素瘦素活性被调节的分子机制,瘦素是一种能参与控制机体食欲和能量消耗的特殊激素;通过对小鼠模型进行研究,研究人员发现,蛋白质Grb10或能促进大脑中瘦素的活性,这或许就为开发基于Grb10的新型疗法来治疗肥胖提供了一定的可能性。
机体控制体重的一种方式就是瘦素,其是由脂肪组织所产生的,机体拥有的脂肪组织越多,就会产生越多瘦素,瘦素会转移到大脑中并且在那里通知专门的神经元机体中所储存的脂肪量,大量的瘦素会告诉大脑中存在大量的脂肪储存,而作为回应,大脑还会诱发抑制食欲并增加能量利用的行为,从而就会导致脂肪组织的减少及体重下降,当一切运行良好时,这种由瘦素介导的脂肪组织和大脑之间的反馈回路就会导致持续性的健康体重。
研究者Yong Xu表示,瘦素能通过抑制食欲同时增强能量消耗来防止体重过度增加的能力就会使其成为一种治疗肥胖的治疗性方法,但很不幸的是,瘦素的补充策略在大多数饮食所诱导的肥胖中被证明是无效的,部分原因或许是由于机体瘦素耐受的产生,这是一种机体中循环的瘦素水平较高的状态,但其并不能抑制机体食物的摄入和体重的增加。
特殊的Grb10蛋白或能提供一种治疗人类肥胖的潜在疗法。
图片来源:Nature Metabolism (2023). DOI:10.1038/s42255-022-00701-x
这项研究中,研究者Xu等人通过研究来寻找能调节瘦素活性以及潜在用来克服机体瘦素耐受的分子,结果发现,蛋白质Grb10或许能作为瘦素活性的一种新型调节子。研究者表示,Grb10能促进瘦素活性的发挥,Grb10的一个特点就是其与此前所发现的瘦素调节子在作用方式上并不相同,Grb10能直接结合神经元上的瘦素受体,从而形成一种复合体;而这种结合就会增强瘦素的信号并帮助减少食物的摄入以及增加能量的消耗;其它的调节子并不能与瘦素受体结合,而是会与其下游的分子所结合。
当消除小鼠大脑中对瘦素反应的神经元中的Grb10时,动物就会摄入更多或者减少能量的消耗并增加体重;从另一方面来讲,增加Grb10的水平或许还会产生一定的好处,其能帮助动物降低对食物的摄入,增加能量消耗并减轻体重;相关研究结果表明,增强Grb10的活性或许就能提供一种方法来增加瘦素信号并帮助减轻体重;本文研究结果支持科学家们进一步调查一种基于Grb10的肥胖疗法的可能性。
目前研究人员非常有兴趣深入调查大脑中Grb10发挥作用的机制,瘦素也能调节机体的情绪和其它情感状态,于是研究人员就想知道是否Grb10也能通过与瘦素受体相互作用来参与调节机体的情绪。综上,本文研究识别出Grb10或许能作为一种潜在的瘦素致敏子,其能通过增强AgRP和POMC神经元对瘦素的反应来维持机体的能量平衡。(生物谷Bioon.com)
原始出处:
Liu, H., He, Y., Bai, J. et al. Hypothalamic Grb10 enhances leptin signalling and promotes weight loss. Nat Metab (2023).doi:10.1038/s42255-022-00701-x
2023-01-23
·生物探索
肥胖已经成为一种全球性“流行病”,现阶段超重/肥胖已经成为严重影响人们身心健康的主要公共卫生问题。近年来,全球超重/肥胖的患病率逐年增长,根据世界卫生组织数据,2025年约五分之一的成年人将患肥胖,我国超重/肥胖的患病率同样不容客观。
根据《中国居民营养与慢性病状况报告(2020年)》显示,我国城乡各年龄段居民超重/肥胖的患病率持续上升,超过二分之一的成年居民患有超重/肥胖,6岁以下、6-17岁的儿童和青少年超重/肥胖的患病率分别为10.4%和19%[1]。
我们知道,肥胖会导致血脂异常、2型糖尿病、高血压等一系列健康问题,同时,其也是部分癌症的影响因素之一。基于此,超重/肥胖俨然已经成为影响我国居民身心健康的主要公共卫生问题。肥胖既是一种慢性疾病状态,也是一种可防可控的慢性非传染性疾病风险因素。
近年来,各国研究人员也正不断研究有效的减重方式。近日,美国贝勒医学院徐勇团队与中南大学湘雅二院刘峰团队合作,通过对小鼠模型进行研究,发现Grb10蛋白可以促进大脑中瘦素(Leptin)的活性,为开发基于Gro10的肥胖治疗新方法奠定了基础。该项研究的结果以“Hypothalamic
Grb10 enhances leptin signalling and promotes weight loss”为题发表在Nature子刊Nature Metabolism上。
图1 研究成果(图源:[2])
Leptin是一种由白色脂肪细胞分泌的激素,它向中枢神经系统传递代谢信息,进而控制能量和葡萄糖稳定,Leptin或Leptin受体(LepR)的功能破坏将导致病态肥胖和糖尿病。下丘脑刺鼠肽基因相关蛋白(AgRP)和阿黑皮素原(POMC)被认为是整合激素和营养信号以维持能量平衡的一级神经元。AgRP和POMC中均有LepR表达,瘦素给药抑制AgRP神经元,激活POMC神经元。
生长因子受体结合蛋白(Grb10)参与调节代谢组织和器官中的多种生物过程,有研究表明其正向调节AgRP和POMC神经元中的瘦素信号,在维持能量稳态中起着关键作用。
在该项研究中,首先,通过免疫荧光试验,研究人员发现87%的AgRP神经元表达Grb10。当AgRP神经元上的Grb10表达下降,会引起正常饮食下小鼠体重增加、能量消耗减少,同时小鼠血清瘦素水平增加。当AgRP神经元上的Grb10过表达,会引起小鼠体重降低、能量消耗增加,同时血清瘦素水平降低。这表明,Grb10在AgRP神经元中通过增加能量消耗阻止体重增加。
图2 AgRP神经元上Grb10表达下降促进肥胖(图源:[2])
为了阐明AgRP神经元中的Grb10是否影响体内瘦素敏感性,研究人员进一步实验。他们发现接受连续3天瘦素治疗的小鼠摄食量明显下降,引起体重降低。而且,Grb10在AgRP神经元中的过表达显著增强了瘦素诱导的厌食症,增加小鼠体重减轻程度。这些结果表明Grb10在AgRP神经元中增强了瘦素敏感性。研究人员还发现,Grb10通过KATP通道的打开增强瘦素抑制AgRP神经元活动能力。
图3 AgRP神经元中的Grb10增强瘦素敏感性(图源:[2])
既往研究表明瘦素可通过诱发ATP敏感的钾离子通道电流抑制AgRP神经元的兴奋,通过电生理实验发现AgRP神经元上的Grb10过表达后明显增强瘦素激活的ATP敏感的钾离子通道,而钾离子通道抑制剂则可以阻断这一兴奋过程。基于此,我们认为,AgRP神经元上的Grb10是通过ATP敏感的钾离子通道,增强瘦素对AgRP神经元的抑制作用,进而起到降低体重的作用。
接着,研究人员发现Grb10在95%的POMC神经元和80%的LepR+ POMC神经元中表达,他们发现当敲除小鼠POMC神经元上的Grb10后,没有引起正常饮食下小鼠的体重及摄食量变化,但却加速高脂饮食小鼠体重增加、脂肪量增加、摄食量增加,血清瘦素和胰岛素水平升高。在通过增加POMC神经元中Grb10的表达后,小鼠体重降低、脂肪量降低、摄食量减少、能量消耗增加,血清瘦素水平下降,糖耐量和胰岛素敏感性改善。
图4 敲除POMC神经元上的Grb10促进肥胖(图源:[2])
对小鼠进行3天的生理盐水或瘦素治疗,以验证Grb10参与POMC神经元的瘦素反应。结果表明,瘦素治疗抑制了小鼠体重增加、摄食量增加,小鼠POMC神经元中Grb10的过表达增强了瘦素在高热量饮食4周后抑制食欲和体重的能力。通过双免疫荧光技术检测,发现POMC神经元中的Grb10有助于在高脂饮食暴露的情况下保留瘦素敏感性。
进一步实验发现,在敲除POMC神经元上的Grb10之后,瘦素引起的瞬时受体电位通道(TrpC)电流减弱,而在POMC神经元上的Grb10过表达之后,TrpC通道电流增强,这表明Grb10通过TrpC增强瘦素对POMC神经元的兴奋性作用。
通过以上研究,发现Grb10是瘦素信号的正向调节因子。Grb10在AgRP神经元中的破坏和过表达分别促进和减少了小鼠的体重增加,这主要是通过改变能量消耗实现的;Grb10在POMC神经元中的过表达可促进小鼠体重减轻。这表明,Grb10在AgRP和POMC神经元中分别以不同的表达机制增强瘦素对食物摄入量和体重的抑制作用。Grb10通过KATP电流放大瘦素诱导的AgRP神经元抑制,并通过TrpC通道激活POMC神经元。
瘦素通过抑制食欲和增加能量消耗以控制体重增加,使其成为潜在的肥胖治疗靶点。由于存在瘦素抵抗作用,循环中的瘦素水平很高,但外源性瘦素不能抑制食欲和体重,故此一直没有得到广泛应用。基于以上研究成果,Grb10可以作为瘦素敏感剂,以增强瘦素对体重的抑制作用,或许可以为减肥治疗带来一种全新的选择。
参考资料:
[1]中国超重/肥胖医学营养治疗指南(2021)[J].中国医学前沿杂志(电子版),2021,13(11):1-55.
[2]Liu H, He Y, Bai J, et al. Hypothalamic Grb10 enhances leptin signalling and promotes weight loss. Nat Metab. 2023 Jan 2. doi: 10.1038/s42255-022-00701-x. Epub ahead of print. PMID: 36593271.
临床前数据
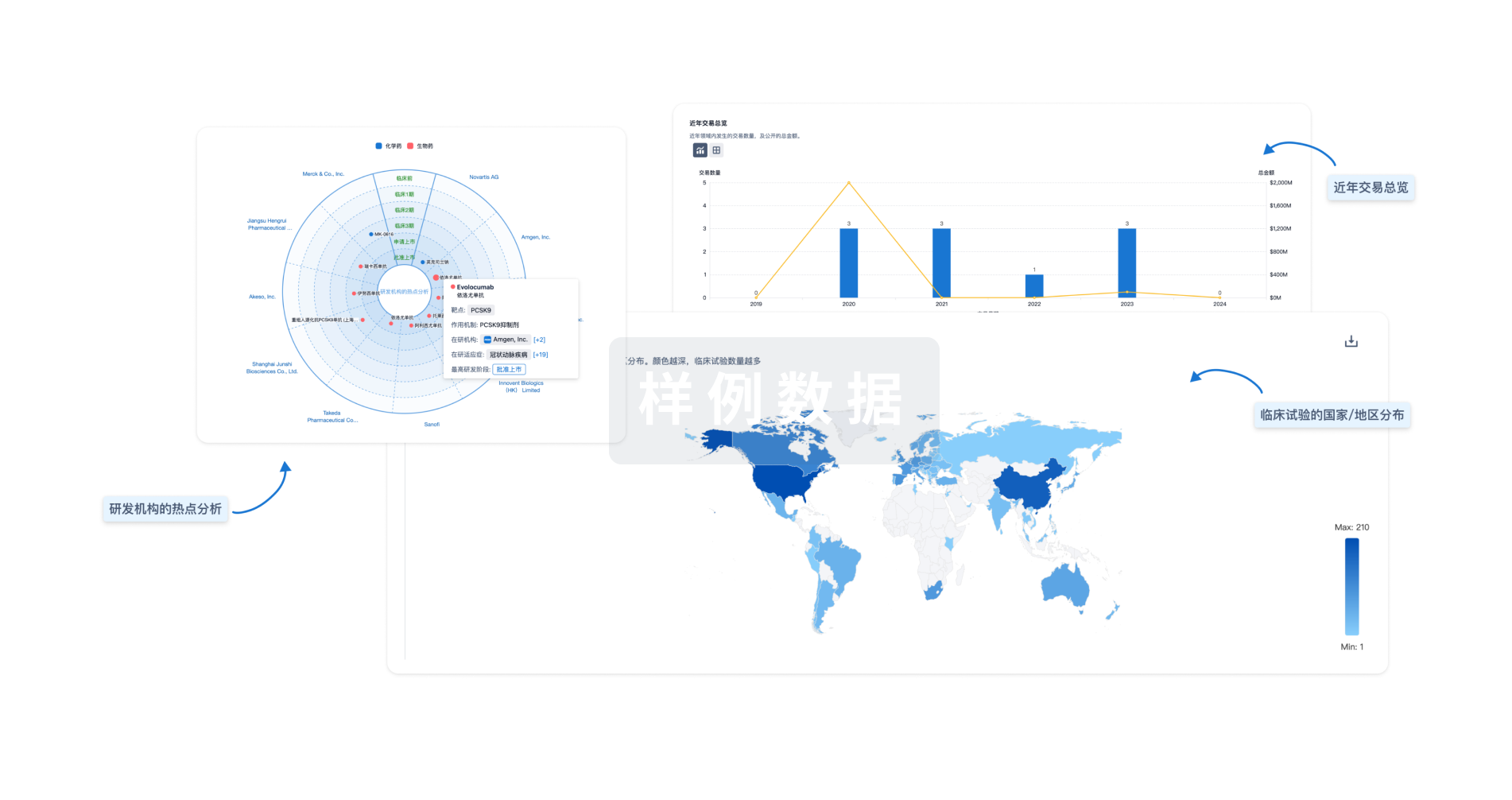
分析
对领域进行一次全面的分析。
登录
或
来和芽仔聊天吧
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用