预约演示
更新于:2025-05-07
Neurotransmitter receptor
更新于:2025-05-07
基本信息
别名 神经递质受体 |
简介- |
关联
7
项与 Neurotransmitter receptor 相关的药物作用机制 神经递质受体调节剂 |
在研适应症 |
非在研适应症- |
最高研发阶段临床3期 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
作用机制 神经递质受体调节剂 |
在研适应症 |
最高研发阶段临床3期 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
作用机制 神经递质受体调节剂 |
在研适应症 |
非在研适应症- |
最高研发阶段临床1期 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
30
项与 Neurotransmitter receptor 相关的临床试验NCT05219617
A Randomized, Double-Blind, Placebo-Controlled Study to Investigate the Efficacy and Safety of Carisbamate (YKP509) As Adjunctive Treatment for Seizures Associated with Lennox-Gastaut Syndrome in Children and Adults, with Optional Open-Label Extension
The primary objective is to evaluate the efficacy of carisbamate (YKP509) as adjunctive treatment in reducing the number of drop seizures (tonic, atonic, and tonic-clonic) compared with placebo in pediatric and adult subjects (age 4-55 years) diagnosed with Lennox Gastaut Syndrome (LGS).
开始日期2022-04-28 |
申办/合作机构 |
NCT04520360
A Single-Center, Open-Label,Randomized, Crossover Study to Evaluate the Relative Bioavailability and Food Effect of a New Oral Suspension and Tablet Formulation of Carisbamate (YKP509) in Healthy Adult Subjects
This study is designed to examine the relative bioavailability of three carisbamate formulations (Oral Suspension Type 1, Oral Suspension Type 2, and a 300 mg Oral Tablet) and to assess the effect of food on the oral bioavailability of the Oral Suspension Type 2 and the 300 mg Oral Tablet.
开始日期2020-08-27 |
申办/合作机构 |
NCT04062981
Phase 1, Open-Label Study of Carisbamate in Adult and Pediatric Subjects With Lennox-Gastaut Syndrome
Open-label extension study from YKP509C001 to evaluate the safety and tolerability of carisbamate in subjects with Lennox-Gastaut Syndrome (LGS).
开始日期2019-05-03 |
申办/合作机构 |
100 项与 Neurotransmitter receptor 相关的临床结果
登录后查看更多信息
100 项与 Neurotransmitter receptor 相关的转化医学
登录后查看更多信息
0 项与 Neurotransmitter receptor 相关的专利(医药)
登录后查看更多信息
1,987
项与 Neurotransmitter receptor 相关的文献(医药)2025-06-01·Neurobiology of Disease
Disrupted synaptic gene expression in Fabry disease: Findings from RNA sequencing
Article
作者: Villacorta-Argüelles, Eduardo ; Leao-Teles, Elisa ; López, Lluis Lis ; Álvarez, J Víctor ; López-Valverde, Laura ; Hermida-Ameijeiras, Álvaro ; Couce, María L ; Vázquez-Mosquera, María E ; Colón-Mejeras, Cristóbal ; López-Mendoza, Manuel ; López-Pardo, Beatriz Martín ; López-Rodríguez, Mónica ; Goicoechea-Diezhandino, María A ; Ortolano, Saida ; Sánchez-Martínez, Rosario ; Guerrero-Márquez, Francisco J
2025-05-01·Neurochemistry International
Astrocyte-neuron metabolic crosstalk in ischaemic stroke
Review
作者: Wang, Qing-Guo ; Lan, Xin ; Chen, Cong-Ai ; Liu, Ying ; Li, Chang-Xiang ; He, Yan-Hui ; Cheng, Jia-Lin ; Cheng, Fa-Feng ; Han, Jin-Hua ; Ren, Zi-Lin ; Wang, Xue-Qian ; Zheng, Yu-Xiao
2025-05-01·JAIDS Journal of Acquired Immune Deficiency Syndromes
Brief Report: Resolution of Neuropsychiatric Adverse Events After Switching to a Doravirine-Based Regimen in the Open-Label Extensions of the DRIVE-AHEAD and DRIVE-FORWARD Trials
Article
作者: Plank, Rebeca M. ; Wan, Hong ; Moyle, Graeme ; Meng, Fanxia ; Sklar, Peter ; Lahoulou, Rima
36
项与 Neurotransmitter receptor 相关的新闻(医药)2025-04-29
脑血管病是一类以脑部血管病变为基础,导致脑组织发生缺血性或出血性损伤的疾病,具有高发病率、高致残率和高死亡率等特点。在全球范围内,脑血管病已成为导致死亡和长期残疾的主要原因之一,给患者及其家庭带来沉重负担。在中国,脑血管病的发病率和患病率呈逐年上升趋势,已成为公共卫生领域亟待解决的重要问题。类器官技术作为生物医学领域的重大突破,近年来发展迅速。类器官是具有一定组织结构和功能的微型器官模型,由干细胞或组织细胞在体外培养形成,能够模拟体内器官的发育、生理和病理过程。自2009年Hans Clevers团队首次成功培育出肠道类器官以来,类器官技术在多个器官系统的研究取得了显著进展。在脑血管疾病领域,类器官技术展现出了广阔的应用前景。通过构建脑类器官,研究人员能够在体外模拟脑血管疾病的发病机制,深入探究血管生成、神经血管单元相互作用等关键环节,为疾病的早期诊断、治疗靶点的发现以及药物研发提供全新的研究平台。尽管类器官技术在脑血管疾病研究领域具有较大潜力,但目前仍面临诸多挑战,其中血管化问题是制约其进一步发展的关键瓶颈。血管化对于类器官的生长、成熟和功能维持至关重要,缺乏完善的血管系统将导致类器官内部营养和氧气供应不足,代谢废物堆积,影响其长期存活和功能发挥。鉴于类器官技术在脑血管疾病研究与治疗领域展现出的潜力以及面临的挑战,本文系统阐述类器官技术在脑血管疾病领域的应用进展,深入探讨血管化脑类器官模型的构建方法,分析基于这些模型的药物研究与疾病机制研究成果,并评估其在脑血管疾病治疗方面的应用前景。1、血管化脑类器官模型的构建体内移植生成血管化脑类器官的首要方法之一是将其植入动物模型中。一项研究采用慢病毒载体标记绿色荧光蛋白(GFP)的人类胚胎干细胞(hESCs)培育大脑类器官胚状体(EBs),并在体外精心培养40~50 d,确保这些类器官具备清晰的胚体边界、径向排列的神经上皮及定义明确的芽。经严格筛选后,这些类器官被移植至成年免疫缺陷小鼠(NOD-SCID)的压后皮质区域。移植手术包括在小鼠大脑中制造一个空腔,将类器官植入其中,并用盖玻片和粘合剂密封,形成颅窗,图1展示了小鼠体内移植类器官模型构建的示意图。▲体内移植类器官模型示意图术后7~10 d,宿主血管开始侵入类器官,术后14 d可形成广泛的血管网络,血管化成功率达到(85.4±6.4)%,且这些血管均源自宿主。这个过程不仅显著提高了类器官的存活率,还促进了神经元的渐进分化和成熟、神经胶质的生成、小胶质细胞的整合,以及轴突向宿主大脑多个区域延伸。通过体内双光子成像技术,研究人员直接观察到移植物中功能性神经元网络和血管的存在。进一步的体内细胞外记录结合光遗传学技术揭示了移植物内神经元的活动,并证实了移植物与宿主大脑之间功能性突触连接的建立。上述发现表明,将脑类器官移植至富含血管的微环境中,可有效利用宿主血管的天然生成能力,实现类器官的血管化和功能化。总体而言,脑类器官的体内移植策略成功克服了坏死核心的限制,通过提供有效灌注显著提升了类器官的活力。体内移植类器官模型在一定程度上弥补了体外类器官模型中血管系统与完整人类大脑生理环境相互作用的缺失,但这一技术目前仍面临诸多问题。首先,模型中的血管成分源自鼠类,这一特点限制了其在人类应用的可移植性,从而阻碍了模型向临床转化的进程。其次,目前观察到的体内移植类器官模型类似于晚期胚胎或早期出生后的组织,其成熟度和空间结构仍与正常血管存在差距。此外,如何精确监测和量化模型的功能表现,尤其是复杂的血流变化、血管壁的应力和变形,仍是技术上的一大难题。体外培养 类器官的内皮化生成血管化脑类器官的关键挑战是协调不同胚层和细胞命运的诱导因子,这些因子常存在相互抑制现象。因此,一些研究方案选择分别培养脑类器官和血管类器官。也有方法通过将非外胚层来源的细胞或其祖细胞直接引入脑类器官中,形成多谱系组装体。一项开创性研究首次尝试生成来源于同一患者的同源结构。该研究利用患者来源的人诱导多能干细胞(hiPSCs)分别生成脑类器官和内皮细胞(ECs),随后在分化后进行共培养。研究中通过无翅型整合位点(Wnt)信号激活诱导中胚层细胞,并采用骨形态发生蛋白4(BMP4)、血管内皮生长因子(VEGF)和成纤维细胞生长因子2(FGF2)分化为内皮祖细胞。在分化至第34天时,将脑类器官重新嵌入含内皮祖细胞的聚合Matrigel基质胶中,继续在体外培养3~5周,最终形成具有较强血管结构的类器官。尽管研究未对神经元区室的细胞结构进行详细描述,但类器官中出现了多条具有穿透性和灌注功能的血管。这些血管表达了人类ECs标志物分化簇31(CD31),证明患者来源的ECs能够成功用于脑类器官的血管化。此后,相关研究将hESCs或hiPSCs与人类脐静脉内皮细胞(HUVECs)共培养,成功构建了具有血管结构的脑类器官模型(vOrganoids)。在培养过程中,细胞首先聚集成胚状体样结构,随后经历神经诱导和分化。研究者发现HUVECs在vOrganoids中形成网状和管状结构,这些结构主要分布于脑室区/亚脑室区上方,而该区域正是神经干细胞和前体细胞的富集地。随着发育进程的推进,血管结构不断向新生神经元区域延伸,且这种状态可持续200 d以上。电生理学进一步检测显示,vOrganoids中存在自发兴奋性突触后电流、自发抑制性突触后电流以及双向电传输现象,表明vOrganoids内部具备化学和电突触。通过单细胞RNA测序分析,研究人员验证了vOrganoids在细胞类型和分子属性上与人类胎儿端脑具有高度相似性,并发现vOrganoids的发育速度相较于非血管化类器官更快。此外,体内移植实验结果证实,vOrganoids可与宿主脑组织实现良好整合,并与宿主血管系统建立了功能性连接。另一类脑类器官血管化策略则是通过基因改造,在脑类器官形成过程中诱导ECs分化。研究显示,从hESCs生成人类皮质类器官,并对其进行基因改造,可异位表达人类ETS突变体2(ETV2)。ETV2能够促使hESCs分化为ECs,这一过程不依赖特定的分化条件,也无需生长因子(如VEGF等)。实验中确定了最佳ETV2表达细胞比例(20%),并于第18天开始诱导ETV2表达,这样处理的类器官可有效形成血管样结构。这些结构不仅在形态上表现为ECs标志物的表达,且在功能上可支持氧气和营养物质的传输,减少细胞死亡,并促进神经元成熟。此外,这些血管化的类器官还展现出血脑屏障(BBB)特性,包括紧密连接、营养转运蛋白和跨内皮电阻表达增加。通过单细胞转录组分析,研究进一步证实了血管化可促进神经元成熟。该模型的一个重要特征是血管状结构被星形胶质细胞和周细胞包围,与人类神经血管单元的组成非常相似。这种单一转录因子介导的方法可跨胚层诱导血管化,并实现类器官中细胞命运诱导时间的精准控制。血管类器官融合作为一种创新性可选方案,一项研究提出了混合神经血管球体新策略,以实现脑类器官的有效血管化。研究者们在hiPSCs基础上独立培养了诱导神经祖细胞(iNPCs)和诱导内皮细胞(iECs)的细胞球体;随后,在两种球体的混合培养基中加入人间充质干细胞(MSC)和SB431542、LDN193189等试剂,形成iNPC-iEC-MSC融合球体。图2展示了体外球体融合类器官模型构建的示意图。▲体外球体融合类器官模型示意图在融合实验中,研究者们精心调整了培养基成分,发现5%的Geltrex基质胶和0.025 wt%的透明质酸能够显著促进球体融合,而高浓度的ROCKi Y27632则对融合过程产生了阻碍效果。通过一系列评估手段,包括细胞增殖、代谢活性检测、细胞因子分泌分析、神经与血管标志物的表达检测以及基因表达分析,研究者们观察到融合后的球体不仅成功促进了神经元和血管细胞的分化,还表现出与脑区特异性标记、基质重塑以及BBB特性相关的基因表达水平显著上调。此外,电生理特性测试进一步证实了融合球体衍生的细胞具备神经元功能和形态学特征。相较于传统的直接混合方法,球体融合策略展现出了显著优势。该策略避免了细胞分离和重新组合的过程,减少了细胞损失,同时允许创建具有精心设计隔间的混合球体结构,从而为研究人员提供了更精准的实验控制。此外,这种精确的排列方式通过组装皮质球体、血管球体和MSC,促进了VEGF-A的分泌,进而加速了皮质组织发育,并展现出层次分布潜力。生物工程技术尽管众多体外培养模型均出现分支血管,且其灌注能力也得到了预测或证实,但这些模型普遍存在一个缺陷,即缺乏主动血流。除此之外,在精确性、重复性、功能模拟以及实验应用等多个方面,也存在不同程度的限制。而生物工程类技术,如3D打印技术、微流控芯片技术等相继涌现与不断发展,为突破这些限制带来了新的希望与可能。微流控芯片技术是在微米尺度上精确操控流体,集成生物、化学、医学分析过程的技术,具有体积小、速度快、用量少等优点,适用于类器官血管化研究中精确控制微环境和细胞共培养。相关研究设计并制造了一种“开放式井”结构的微流控芯片,利用3D打印技术,并选用Dental SG树脂作为打印材料。通过特殊的清洗程序,确保了芯片的生物相容性。该芯片中央是类器官培养室,两侧设有微流控通道,二者通过50 μm的间隙相连,允许细胞和分子交换。血管细胞和脑类器官均从人类多能干细胞(hPSC)分化而来。在分化的第6天,血管细胞被接种至微流控通道中;而在分化的第5天,脑类器官则被接种至类器官培养室,实现了二者的共培养。在含有VEGF-A培养基的诱导下,血管细胞从微流控通道向类器官培养室伸出血管芽,形成有序的血管网络,并与脑类器官相互作用,最终形成整合的神经血管类器官。共培养实验结果表明,血管细胞的存在可能加速脑类器官的成熟过程。此外,生成的血管网络不仅结构完整,且具有功能性,可进行小分子灌注和通透性实验。相较于体内移植模型,各种体外类器官模型具有环境变量控制性强、灵活性和重复性高、伦理问题少等一系列优势。但体外模型无法完全再现体内复杂的生理和生物学条件、无法模拟真实的血液动力学环境,虽可通过培养不同类型的细胞模拟细胞群体和细胞间的交互作用,但其行为可能与体内复杂的组织环境不同,导致实验结果出现偏差。对于长时间体外培养的血管化脑类器官,如何保证组织的持续生长和功能稳定也是面临的一项技术难题。各种生物工程技术展现出了解决上述问题的潜力,但现阶段在应用方面仍不够成熟,需进一步深入研究。2、基于血管化脑类器官的药物研发BBB是维持中枢神经系统稳态的关键屏障,其通透性调控直接关系到药物跨屏障输送的效率以及在脑组织中的疗效,是中枢神经系统药物研究不可忽视的重要因素。血管化脑类器官在一定程度上再现了BBB的结构与功能,为中枢神经系统药物研发提供了一个全新平台。Saglam-Metiner等介绍了一种ICU患者芯片平台,该平台将hiPSCs分化的肥大细胞与脑类器官相结合,并在三维基质中进行培养,从而成功模拟了BBB与脑组织之间的相互作用。该平台利用一层覆盖有人类脑微血管ECs的膜模拟血管腔与组织室的分离,部分再现了BBB的结构和功能。研究发现,异丙酚能够激活肥大细胞、微胶质细胞和星形胶质细胞,进而增加一系列促炎基因的表达。同时,还伴随脑组织中缝隙连接蛋白、抑制性神经递质受体以及兴奋性神经递质受体表达水平升高。表明异丙酚可能通过激发神经炎症反应和改变神经递质平衡,进而影响神经元的功能和存活。相比之下,咪达唑仑对BBB的破坏作用更为显著,其能够降低ECs之间的电阻值、增加屏障通透性及损伤屏障完整性。此外,咪达唑仑还可激活肥大细胞和小胶质细胞,加剧谷氨酸相关的神经毒性,并诱导白细胞介素、干扰素等炎症因子过度表达,这些作用最终导致脑组织细胞凋亡明显增多。咪达唑仑还抑制了抑制性神经递质受体和兴奋性神经递质受体的表达,进一步扰乱神经递质平衡,从而对脑组织功能造成显著负面影响。与传统研究模型相比,血管化脑类器官技术结合微流控平台更贴近人脑的生理环境,可更准确地反映药物在脑部的作用机制,为中枢神经系统药物研究提供了强有力工具,有助于推动相关药物研发工作的深入开展。但BBB并非孤立存在,而是由ECs、星形胶质细胞、平滑肌细胞等共同作用维持,模拟这些周围细胞与ECs相互作用对于再现BBB功能同样重要,如何通过体外模型进一步复现BBB紧密连接的结构和选择性透过药物分子的功能是后续研究的重点。3、脑血管疾病机制的探究脑类器官模型在模拟人体组织三维结构及细胞间复杂相互作用方面更具精准性,为深入探究疾病发病机制带来了独特优势。近期,一项研究借助脑类器官芯片,为遗传性出血性毛细血管扩张症(HHT)1型发病机制的研究提供了新的视角与见解。HHT是一种以血管脆弱为显著特征的遗传性疾病,其中HHT 1型是由ENG(ENDOGLIN)基因突变所引发。该研究从1例罕见的嵌合HHT 1型患者体内获取细胞,成功生成了同源的突变型和正常型hiPSCs,并进一步将其分化为ECs,从而在体外模拟出HHT 1型血管状况。研究结果显示,在二维培养条件下,突变型和正常型ECs的功能表现相似。然而,当其置于三维模型中时,突变型ECs所形成的血管网络却呈现出诸多组织缺陷:血管密度明显降低、直径变小、ECs数量减少,且血管渗漏现象显著增加。此外,突变型ECs与脑血管周细胞之间的相互作用也受到损害,周细胞的覆盖率大幅下降,且与ECs的距离更远,进一步加剧了血管的脆弱性。荧光葡聚糖渗漏实验进行验证,证实突变型血管的渗漏程度更高,反映出其屏障功能出现明显受损。此外,另一项研究通过构建患者源性类器官,成功复制了脑海绵状畸形(CCM)的特征,为深入理解CCM的发病机制提供了重要见解。CCM是一种遗传性脑血管疾病,其特征是由众多薄壁血管组成的海绵状异常血管团,其易破裂出血,导致卒中和神经功能障碍。该研究观察到,CCM患者源性的BBB类器官中内皮通道异常扩大,且通道呈现背靠背排列,这一现象与体内CCM的病理特征高度相似。进一步分析显示,这些类器官中紧密连接蛋白和基底膜蛋白的表达显著降低,这表明BBB的完整性受到了破坏。单细胞RNA测序揭示了ECs中病变标记基因显著上调,以及血管生成相关基因表达增强,表明ECs的病理变化在CCM发病机制中占据主导地位。此外,研究还发现CCM患者的MSC向血管平滑肌细胞分化的潜能减弱,这种平滑肌细胞的发育损失可能是导致CCM发病的关键因素之一。4、脑血管疾病治疗的新策略类器官技术作为一种新兴的脑血管疾病治疗策略,正展现出较大发展潜力。2020年,一项研究首次探索了脑类器官技术在缺血性卒中治疗中的可行性和潜在机制。研究人员将培养了55 d的脑类器官移植至大鼠大脑中动脉闭塞模型中,发现脑类器官移植后不仅显著减小了脑梗死体积,还显著改善了神经功能,这种效果与脑类器官的多系分化能力密切相关。移植的脑类器官能够模拟体内皮质发育,分化出多种细胞类型,包括神经前体细胞、神经元和星形胶质细胞等,通过神经发生、突触重建、轴突再生和血管生成作用,促进脑损伤修复。此外,移植细胞可沿胼胝体广泛迁移并在受损区域长时间存活。研究还表明,卒中后6 h或24 h内移植类器官可显著减轻脑损伤并改善行为表现,而超过7 d则移植效果有限,这一研究为脑类器官作为一种新型缺血性卒中治疗策略提供了重要证据。随后,另一项研究在该领域进一步探索,尤其是脑类器官在长期组织修复和功能恢复中的作用。研究人员利用hPSC诱导生成脑类器官,并将其移植至NOD-SCID小鼠脑梗死核心与梗死周边交界处,数月后类器官在脑梗死核心区域内可长期存活,分化为目标神经元,形成与宿主神经网络相连接的功能性突触,恢复受损脑组织结构。移植的类器官在梗死区域内分化为成熟的谷氨酸能神经元,还通过轴突延伸连接至远端脑区,整合至宿主神经回路。这种结构修复和功能整合体现在小鼠行为改善上,包括感知和运动功能的恢复。与单一细胞移植相比,脑类器官移植表现出更高的细胞存活率、更强的组织修复能力和更优越的神经功能恢复效果。这一研究完善了脑类器官移植的实施方法,证明其可重建梗死组织并恢复卒中后功能障碍。由此可见,脑类器官为脑血管疾病的治疗开辟了一条极具潜力的新途径,其在缺血性卒中治疗领域的应用前景令人期待。5、小结与展望类器官技术为脑血管疾病的研究和治疗开辟了新视角,其在疾病机制探究、药物研发及治疗策略中的应用展现出了广阔潜力。通过体内移植、体外培养以及生物工程技术的创新发展,血管化脑类器官的构建逐步克服了传统模型在结构和功能上的局限,为研究神经血管单元相互作用、血管生成及BBB功能提供了更接近人体生理环境的实验平台。此外,这些模型在药物筛选中实现了中枢神经系统药物效应的精确评估,为复杂脑血管疾病的发病机制研究提供了突破性手段,在治疗策略方面展现出了重要潜力。尽管类器官技术取得了诸多进展,但仍面临诸多挑战。伦理问题和安全性评估是其临床应用中的关键问题,特别是涉及人类干细胞问题。人类干细胞的使用(特别是胚胎干细胞),涉及伦理问题,因其来源于胚胎,且在胚胎发育过程中可能导致生命终结。为确保其伦理合规性,所有使用人类干细胞的研究均必须严格遵守国际伦理规范,确保研究参与者知情同意,并保证数据的透明性。在安全性评估方面,类器官技术的临床转化面临细胞异质性、免疫反应、肿瘤发生风险等挑战。类器官在体外培养中的异质性可能导致不同患者之间的效果差异。因此,需通过严格的动物实验和长时间的安全性测试评估其临床应用风险。此外,类器官移植后可能引发免疫排斥反应、癌变风险等问题,需通过实验数据进一步验证,并制定详细的安全性评估标准。在技术实现方面,体外类器官模型在控制环境变量和伦理问题方面具有优势,但难以完全再现体内复杂的生理环境和血液动力学,且长期培养时如何维持功能稳定仍是难题。体内移植模型虽然弥补了这一缺陷,但血管成分源自鼠类,限制了其在人体的临床应用。此外,这些模型的成熟度和血管功能尚存在差距,血管化效率和主动灌注能力的进一步优化亟待解决。BBB的模拟也面临挑战,特别是如何有效复现紧密连接和药物透过性方面,需开展进一步研究和技术突破。结合微流控技术及高通量3D打印等前沿手段,未来有望进一步提升类器官模型的精确性和功能性。特别是在脑血管疾病领域,探索血管化脑类器官在个性化治疗方面的应用潜力,如针对特定患者设计定制化的药物筛选平台和治疗方案,将推动精准医疗的发展。与此同时,随着生物工程技术的进步和多学科协作的深化,类器官技术有望逐步实现从实验室到临床的转化,为脑血管疾病的诊疗带来革命性突破。相关阅读中风“治愈率”提高53%!浙附二院楼敏团队研究登NEJM:这些患者治疗“时间窗”延长至24小时!脑萎缩速度下降57%,认知功能改善!《自然-医学》:干细胞疗法为阿尔茨海默病带来治疗希望心梗前35年或已出现!年轻血脂高,增加这些心脑血管疾病风险欢迎投稿:学术成果、前沿进展、临床干货等主题均可,点此了解投稿详情。参考资料(可上下滑动查看)[1]Walter K. What is acute ischemic stroke?[J]. JAMA,2022,327(9): 885.[2]Montaño A,Hanley D F,Hemphill J C,3rd. Hemorrhagic stroke[J]. Handb Clin Neurol,2021,176: 229-248.[3]Goldstein L B. Introduction for focused updates in cerebrovascular disease[J]. Stroke,2020,51(3): 708-710.[4]Li Y,Zeng P M,Wu J,et al. Advances and applications of brain organoids[J]. Neurosci Bull,2023,39(11): 1703-1716.[5]Sato T,Vries R G,Snippert H J,et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche[J]. Nature,2009,459(7244): 262-265.[6]Song L Q,Yan Y W,Marzano M,et al. Studying heterotypic cell-cell interactions in the human brain using pluripotent stem cell models for neurodegeneration[J]. Cells,2019,8(4): 299.[7]Orlova V V,Nahon D M,Cochrane A,et al. Vascular defects associated with hereditary hemorrhagic telangiectasia revealed in patient-derived isogenic iPSCs in 3D vessels on chip[J]. Stem Cell Reports,2022,17(7): 1536-1545.[8]Saglam-Metiner P,Yanasik S,Odabasi Y C,et al. ICU patient-on-a-chip emulating orchestration of mast cells and cerebral organoids in neuroinflammation[J]. Commun Biol,2024,7(1): 1627.[9]Hofer M,Lutolf M P. Engineering organoids[J]. Nat Rev Mater,2021,6(5): 402-420.[10]Mansour A A,Gonçalves J T,Bloyd C W,et al. An in vivo model of functional and vascularized human brain organoids[J]. Nat Biotechnol,2018,36(5): 432-441.[11]Wilson M N,Thunemann M,Liu X,et al. Multimodal monitoring of human cortical organoids implanted in mice reveal functional connection with visual cortex[J]. Nat Commun,2022,13(1): 7945.[12]Pham M T,Pollock K M,Rose M D,et al. Generation of human vascularized brain organoids[J]. Neuroreport,2018,29(7): 588-593.[13]Kistemaker L,van Bodegraven EJ,de Vries HE,Hol EM. Vascularized human brain organoids: current possibilities and prospects[J]. Trends Biotechnol,2025,S0167-7799(24)00357-3.doi: 10.1016/j.tibtech.2024.11.021. Epub ahead of print.[14]Cakir B,Xiang Y F,Tanaka Y,et al. Engineering of human brain organoids with a functional vascular-like system[J]. Nat Methods,2019,16(11): 1169-1175.[15]Wörsdörfer P,Dalda N,Kern A,et al. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells[J]. Sci Rep,2019,9(1): 15663.[16]Shi Y C,Sun L,Wang M D,et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo[J]. PLoS Biol,2020,18(5): e3000705.[17]Song L Q,Yuan X G,Jones Z,et al. Assembly of human stem cell-derived cortical spheroids and vascular spheroids to model 3-D brain-like tissues[J]. Sci Rep,2019,9(1): 5977.[18]Tan S Y,Feng X H,Cheng L K W,et al. Vascularized human brain organoid on-chip[J]. Lab Chip,2023,23(12): 2693-2709.[19]Salmon I,Grebenyuk S,Abdel Fattah A R,et al. Engineering neurovascular organoids with 3D printed microfluidic chips[J]. Lab Chip,2022,22(8): 1615-1629.[20]Xu C,Alameri A,Leong W,et al. Multiscale engineering of brain organoids for disease modeling[J]. Adv Drug Deliv Rev,2024,210: 115344.[21]Dao L,You Z,Lu L,et al. Modeling blood-brain barrier formation and cerebral cavernous malformations in human PSC-derived organoids[J]. Cell Stem Cell,2024,31(6): 818-833.e11.[22]Wang S N,Wang Z,Xu T Y,et al. Cerebral organoids repair ischemic stroke brain injury[J]. Transl Stroke Res,2020,11(5): 983-1000.[23]Cao S Y,Yang D,Huang Z Q,et al. Cerebral organoids transplantation repairs infarcted cortex and restores impaired function after stroke[J]. NPJ Regen Med,2023,8(1): 27.[24]Zheng F Y,Xiao Y M H,Liu H,et al. Patient-specific organoid and organ-on-a-chip: 3D cell-culture meets 3D printing and numerical simulation[J]. Adv Biol (Weinh),2021,5(6): e2000024.免责声明:本文仅作信息交流之目的,文中观点不代表药明康德立场,亦不代表药明康德支持或反对文中观点。本文也不是治疗方案推荐。如需获得治疗方案指导,请前往正规医院就诊。分享,点赞,在看,传递医学新知
2025-04-17
·动脉网
动脉网第一时间获悉,近日,全球首家神经递质分泌内源性调控创新药研发企业安然菲(昆山)新药研发有限公司(下称“安然菲”)宣布完成数千万元人民币天使轮融资,本轮融资由峰瑞资本独家投资,格物资本担任独家财务顾问。本轮资金将主要用于以帕金森病为代表的多种神经系统疾病First-In-Class创新药管线研发。安然菲成立于2023年,创始人张燕峰博士为英国埃克塞特大学神经科学教授,牛津大学客座讲师,有超过十五年对于多巴胺分泌及功能的研究经历,并在Nature Neuroscience、Neuron、Nature Communications等国际顶级期刊发表多篇重要学术论文并担任审稿人。创始人所在的牛津大学帕金森病研究中心(OPDC)是全世界综合实力最强的跨学科帕金森病研究中心。安然菲致力于研发调控神经递质分泌的同类最先、同类最优的创新药,未来将会在全球范围内解决一系列未得到满足的临床需求,推动神经科学领域的治疗变革,造福广大患者。01神经递质分泌失衡引发多种神经类疾病非正常的神经递质释放已证实与多种神经疾病密切相关,如多巴胺分泌不足会引发帕金森症,多动症,成瘾症,精神分裂症,暴食症等。截至2021年,全球神经精神类疾病患者超过4.7亿,相关药物销售额超300亿美元。其中,帕金森患者全球超过700万,中国患者约310万,是第二大神经退行性疾病,全球帕金森相关药物年销售额超过400亿人民币。02现有疗法为外源性调节神经递质浓度,存在明显副作用目前药物治疗仍是中枢神经系统疾病的首选,一般以神经递质及其受体作为靶点成药,包括外源性增加神经递质浓度,结合神经递质受体产生激活或者抑制效果等等。然而,现有方法往往使得大脑环路中,下游神经元接收到的信号并不是由上游神经元编码,而来自于由药物浓度决定的非生理性信号。虽然这些信号可以很强,但长期且非生理性的刺激会给下游神经环路带来明显的副作用。譬如目前治疗帕金森症的唯一的“金标准”药物左旋多巴,可以在脑内转变为多巴胺,弥补帕金森患者纹状体内缺乏的这类神经递质。但长期服用左旋多巴后,患者会出现严重的运动障碍(异动症)和其它精神类并发症,以至于左旋多巴的有效期仅为5年。03全球首创神经递质内源性调节技术平台安然菲建立了全球首创的技术平台。该平台通过调节神经细胞的电信号到化学信号的转化效率,可以在不影响神经细胞放电频率即不改变其生理意义的前提下,通过内源性地调节神经递质分泌,直接对目标神经递质分泌进行生理性调控进行针对各种疾病的治疗。该方法无需侵入性手术,而且相对传统疗法具有诸多优势:1)相对于传统疗法的持续不变的的高强度干扰神经间的信号传导,该治疗为生理性调节,不改变神经细胞放电的原有频率,从而避免了因长期激活下游细胞而造成的功能损伤。2)治疗后,疾病受损的神经间的信号将得到相应的放大(或缩小)。该治疗确保了细胞间的沟通恢复成正常的水平,在保证疗效的同时,进一步降低了副作用。3)由于该治疗针对性强,治疗目标准确,相对于传统疗法,新平台能够大幅减少因为对其他种类神经信号干扰而引起的副作用。以上特性使得这种全球首创的新型疗法适用多种疾病,可解决一系列未得到满足的临床需求。04首个管线瞄准帕金森症First-In-Class创新药研发,研究成果已在国际顶刊NATURE NEUROSCIENCE上发表据悉,该公司首个管线瞄准帕金森症First-In-Class创新药,全新的靶点首次实现了对多巴胺能神经元分泌的直接调控,该疗法采用的全新机理颠覆对多巴胺分泌机制的认知,成药后可单独使用治疗帕金森症(不依赖左旋多巴)。目前临床前研究数据证明其单独治疗可以显著提高多巴胺分泌,经治疗后的帕金森模型小鼠均恢复了运动功能。相关研究成果已在权威学术期刊NATURE NEUROSCIENCE上发表,标志着相关的神经生物学机制已获得学术界认可。基于该技术平台,安然菲首先针对帕金森开展了相关药物管线布局。据悉,新靶点实现了对多巴胺能神经元分泌的直接调控,成药后可单独针对帕金森症进行治疗。相关研究成果已在NATURE NEUROSCIENCE上发表。05资本青睐,安然菲有望快速进入临床研究阶段,新一轮融资同步开展目前,安然菲已完成先导化合物等多项国际专利申请,针对先导化合物的优化工作即将完成,预计2025年确认临床前候选化合物(PCC)。对于本次融资,安然菲CEO张燕峰表示,感谢峰瑞资本对公司的支持,资本加持下公司将于近期完成首个管线帕金森创新药的PCC确认,及以快速扫描循环伏安法(FCV)为代表的神经电生理平台的拓展,未来还将基于内源性调控多巴胺新机制的技术平台拓展多动症、抽动症等多管线研发。峰瑞资本谢达博士表示,中枢神经系统疾病存在着巨大的临床未满足需求,然而受限于复杂的脑疾病病理,以及局限的研究手段和研究样本,神经精神疾病药物的开发仍然面临巨大的挑战。安然菲立足于全新的神经生物学机制开发中枢神经系统疾病药物,关注对脑局部网络的调控,有别于过往单一的递质回补或对递质受体的直接干预,因而有望获得更贴近生理水平的治疗效果,并具备在不同类型的神经元、不同物种上拓展和验证的潜力。通过提高大脑内多巴胺浓度治疗帕金森病已经得到临床认可,安然菲的首条管线也将专注帕金森病,以期为新的生物学机制提供关键临床验证,为帕金森患者带来更优质的药物。格物资本合伙人邸海明表示,非常荣幸能够和全球帕金森病研究领域的顶级科学家合作,张博在电生理,电化学,药理学,行为学,化学,和计算机模拟均具有深厚的背景,安然菲在研的用于治疗帕金森的首个管线机理清晰、临床前研究数据已展现出优异的效果,未来有望打破左旋多巴恐惧症,为帕金森患者带来新的福音。公司新一轮融资也正式开始,期待与更多投资人交流。如果您想对接文章中提到的项目,或您的项目想被动脉网报道,或者发布融资新闻,请与我们联系;也可加入动脉网行业社群,结交更多志同道合的好友。近期推荐声明:动脉网所刊载内容之知识产权为动脉网及相关权利人专属所有或持有。未经许可,禁止进行转载、摘编、复制及建立镜像等任何使用。动脉网,未来医疗服务平台
2025-03-29
点击蓝字关注我们本周,热点较多。首先看审评审批方面,国内外都有两个重磅药获批,国内来说,科睿药业抗流感1类新药玛舒拉沙韦获批上市,国外而言,first-in-class口服抗生素获FDA批准上市,成为近30年来首个获批治疗uUTI的新型口服抗生素。其次看研发方面,多个药物取得重要进展,值得一提的就是,中美瑞康公布小核酸癌症新药积极结果,完全缓解率超66%,但比较遗憾的是,Cassava Sciences阿尔茨海默病新药Ⅲ期研究失败;最后是交易及投融资方面,比较值得关注的就是,默沙东近20亿美元引进恒瑞Lp(a)抑制剂。本期盘点包括审评审批、研发以及交易及投融资三大板块,统计时间为2025.3.24-3.28,包含25条信息。 审评审批NMPA上市批准1、3月27日,NMPA官网显示,青峰医药下属子公司科睿药业1类新药玛舒拉沙韦片(GP681片)获批上市,用于既往健康的12岁及以上青少年和成人单纯性甲型和乙型流感患者的治疗,不包括存在流感相关并发症高风险的患者。玛舒拉沙韦片是青峰医药与银杏树药业联合开发的一种聚合酶酸性蛋白(PA)抑制剂,患者全病程只需服药一次。申请2、3月24日,CDE官网显示,Plethora Solutions和复星医药共同申报的5.1类新药利多卡因丙胺卡因气雾剂(研发代号:PSD502)申报上市,用于治疗早泄。这是一款利多卡因及丙胺卡因的专有配方,为一款定量透皮喷雾,最早于2013年11月在欧盟获批上市,2018年12月,复星医药控股子公司万邦生化医药获得该产品在中国商业化权益。3、3月25日,CDE官网显示,施维雅申报的5.1类新药艾伏尼布片新适应症申报上市,推测适应症为联合阿扎胞甘一线治疗携带易感IDH1突变的AML患者。艾伏尼布片是一种针对IDH1突变酶的口服靶向抑制剂,此前已作为“临床急需境外新药”在国内获批,用于治疗携带IDH1易感突变的复发或难治性急性髓系白血病(AML)成人患者。临床批准 4、3月24日,CDE官网显示,恩华药业1类新药NH140068片获批临床,拟开发治疗精神分裂症。NH140068是一款适于口服的多种神经递质受体激动剂,为新一代治疗精神分裂症的创新药。本次是该产品首次在中国获批临床。5、3月25日,CDE官网显示,科伦博泰1类新药SKB107注射液获批临床,拟开发用于晚期实体瘤骨转移。这是该公司与西南医科大学附属医院共同开发的靶向肿瘤骨转移的放射性核素偶联药物(RDC)药物。本次是该产品首次在中国获批IND。6、3月25日,CDE官网显示,映恩生物1类新药注射用DB-2304获批临床,拟开发治疗系统性红斑狼疮。DB-2304是映恩生物自主研发的靶向BDCA2这一浆细胞样树突状细胞(pDC)特异靶点的ADC产品,具有潜在“first-in-class”。7、3月25日,CDE官网显示,复宏汉霖的HLX79注射液获批Ⅱ期临床,联合利妥昔单抗(汉利康)治疗活动期肾小球肾炎。HLX79是复宏汉霖许可引进的一款潜在“first-in-class”人唾液酸酶融合蛋白,2024年12月,复宏汉霖与Palleon达成一项9530万美元的授权合作,获得HLX79在中国的独家许可。8、3月27日,CDE官网显示,华东医药1类新药HDM3019获批临床,拟开发治疗类风湿关节炎。HDM3019是靶向OX40L和TNFα的双特异性抗体。华东医药于2024年8月与IMBiologics达成独家许可协议,获得两款自身免疫性疾病新药在中国等37个亚洲国家的权益,其中就包括了这款IMB-101。该项合作的总金额超3亿美元。9、3月27日,CDE官网显示,阿斯利康旗下Alexion的1类新药Eneboparatide注射液获批临床,拟开发治疗慢性甲状旁腺功能减退症。这是一款治疗性多肽,2024年3月,阿斯利康宣布以10.5亿美元收购罕见内分泌疾病领域生物技术公司Amolyt Pharma,从而获得这款名为eneboparatide(AZP-3601)的Ⅲ期临床阶段研发项目。10、3月27日,CDE官网显示,礼来申报的1类新药注射用LY4052031获批临床,拟单药用于治疗晚期或转移性尿路上皮癌或其他实体瘤。这是一款在研的抗Nectin-4抗体偶联药物(ADC),正在国际范围内开展Ⅰ期临床研究。本次是该产品首次在中国获批IND。优先审评11、3月28日,CDE官网显示,默沙东的Clesrovimab注射液拟纳入优先审评,用于即将进入或出生在第一个呼吸道合胞病毒(RSV)流行季的新生儿和婴儿预防RSV所致的下呼吸道感染。Clesrovimab是一种在研的半衰期延长的单抗,可作为被动免疫手段,用于预防RSV疾病。2024年12月,该产品向FDA批准上市,用于保护婴儿在其首个RSV季节免受RSV疾病的侵害。FDA预定在2025年6月前完成审评。突破性疗法12、3月27日,CDE官网显示,恒瑞医药1类新药HRS-5965胶囊拟纳入突破性治疗品种,适应症为原发性IgA肾病。HRS-5965为一款补体因子B抑制剂。该产品针对原发性IgA肾病适应症目前正处于Ⅱ期临床阶段。HRS-5965的首个Ⅲ期临床正在进行中,旨在评估HRS-5965胶囊对比一款C5补体抑制剂治疗阵发性睡眠性血红蛋白尿(PNH)的有效性与安全性。FDA上市批准13、3月26日,FDA官网显示,GSK的Gepotidacin(商品名:Blujepa)获批上市,用于治疗单纯性尿路感染(uUTI)的成人及12岁以上青少年患者。Gepotidacin是一款具有独特作用机制的口服“first-in-class”抗生素。14、3月26日,FDA官网显示,Exelixis的卡博替尼新适应症获批,用于治疗既往接受过治疗、不可切除的、局部晚期或转移性的、分化良好的胰腺神经内分泌肿瘤(pNET)和胰腺外NET(epNET)儿童(≥12岁)和成人患者。这是第一个获FDA批准用于系统治疗无论原发肿瘤部位、分级、生长抑素受体表达和功能状态的经治NET的药物,于2012年11月在美国获批上市。临床批准15、3月24日,FDA官网显示,神济昌华的SNUG01获批Ⅰ/Ⅱa期国际多中心注册临床试验,用于治疗肌萎缩侧索硬化(ALS)。SNUG01是以重组腺相关病毒9型(rAAV9)为载体,通过鞘内注射(IT)的方式,将人源TRIM72基因靶向递送至神经元。16、3月24日,FDA官网显示,云顶新耀的EVM14注射液获批临床。EVM14是一款靶向多种肿瘤相关抗原(TAA)的通用型的现货肿瘤治疗性疫苗,拟用于非小细胞肺癌、头颈癌等多种癌症的治疗。该产品通过将编码多种肿瘤相关抗原的mRNA原液包封在脂质系统中配制而成。优先审评17、3月25日,FDA官网显示,赛诺菲的Tolebrutinib获优先审评,用于治疗非复发性继发进展型多发性硬化症(nrSPMS)并延缓成人患者独立于复发活动的残疾进展,PDUFA日期为2025年9月28日。欧盟也正在审评该药的上市申请。如果获得批准,Tolebrutinib将成为第一种也是唯一一款既能治疗nrSPMS又能减缓残疾累积而不受复发活动影响的脑穿透性BTK抑制剂。研发临床状态18、3月24日,索元生物宣布,其DB107治疗新诊断的高级别胶质瘤的临床Ⅰ/Ⅱ期试验首例受试者完成入组。这是一项评估DB107在切除时和术后静脉注射,联合DB107FC和放射治疗或DB107FC、替莫唑胺(TMZ)和放射治疗对新诊断的高级别胶质瘤患者进行治疗的研究,计划招募70名患者,证明DB107在生物标志物阳性脑瘤患者中无进展生存率的改善。临床数据19、3月24日,Opthea Limited公布了Sozinibercept治疗湿性年龄相关性黄斑变性(wAMD)的Ⅲ期COAST研究。结果显示,该研究未达到主要终点。在总体人群中,Sozinibercept(每4周1次)联合阿柏西普组、Sozinibercept(每8周1次)联合阿柏西普组和安慰剂联合阿柏西普组的BCVA评分分别增加了13.5个字母、12.8个字母和13.7个字母,组间p值分别为0.86和0.42。Sozinibercept是一款靶向血管内皮生长因子受体3(VEGFR3)的Fc融合蛋白,可阻断VEGF-C、VEGF-D与VEGFR2、VEGFR3的结合。20、3月24日,中美瑞康公布了RAG-01治疗非肌层浸润性膀胱癌(NMIBC)Ⅰ期临床试验的积极初步数据。数据显示,在接受卡介苗(BCG)治疗失败的患者中,最低两个剂量组的原位癌(CIS)患者完全缓解率(CR)达到66.7%,且安全性表现优异。RAG-01是一种创新性的saRNA(小激活RNA)疗法,旨在上调p21肿瘤抑制基因的表达。21、3月25日,Cassava Sciences公布了Simufilam治疗轻度至中度阿尔茨海默病(AD)的Ⅲ期REFOCUS-ALZ研究数据。数据显示,未达到主要终点。此前,该研究因Ⅲ期RETHINK-ALZ研究失败在2024年11月25日终止开展。Simufilam是一款靶向错构的细丝蛋白A(FLNA)的口服小分子药物。22、3月28日,劲方医药公布了氟泽雷塞联合西妥昔单抗一线治疗NSCLC的最新Ⅱ期(KROCUS)数据。数据显示,该药具有优秀的总体疗效、显著的脑转移患者肿瘤缓解、以及优于二线及以上氟泽雷塞单药疗法的安全性/耐受性;相较于包含免疫疗法的当前一线NSCLC标准治疗,KROCUS方案也提示了针对KRAS突变患者的更优治疗潜力。氟泽雷塞为一款KRAS G12C抑制剂,由信达生物和劲方医药合作开发,是中国批准上市的首个KRAS抑制剂药物。交易及投融资23、3月24日,联邦制药宣布,其全资附属公司联邦生物已与诺和诺德签订协议。诺和诺德将获得UBT251全球(不包括中国大陆、中国香港、中国澳门和中国台湾地区)的开发、生产和商业化权益。联邦生物保留中国权益。UBT251是一款长效GLP-1R/GIPR/GCGR三重激动剂,拟开发用于治疗肥胖症、2型糖尿病和其他疾病。根据协议,联邦生物将获得2亿美元首付款和最高18亿美元的潜在里程碑付款,以及分层销售提成。24、3月25日,恒瑞医药宣布,与默沙东就其脂蛋白(a)[Lp(a)]口服小分子项目(包括名为HRS-5346先导化合物)达成独家许可协议。根据协议,恒瑞医药将HRS-5346在大中华区以外的全球范围内开发、生产和商业化的独家权利有偿许可给默沙东。恒瑞医药将收取2亿美元的首付款,并有资格获得不超过17.7亿美元的与特定的开发、监管和商业化相关的里程碑付款,以及如果相关产品获批上市,基于HRS-5346的净销售额的销售提成。25、3月26日,浦合医药宣布,与拜耳就BAY 3713372(浦合原代号PH020)达成全球许可协议。根据协议,拜耳获得开发、制造和商业化该产品的全球独家许可。BAY 3713372是一种口服的强效选择性MTA协同PRMT5抑制剂,拜耳已招募首例患者参与Ⅰ期人体首次剂量爬坡临床试验,研究该产品用于治疗MTAP缺失型实体瘤。END2025金笔奖征文活动开启,来投稿吧!领取CPHI & PMEC China 2025展会门票来源:CPHI制药在线声明:本文仅代表作者观点,并不代表制药在线立场。本网站内容仅出于传递更多信息之目的。如需转载,请务必注明文章来源和作者。投稿邮箱:Kelly.Xiao@imsinoexpo.com▼更多制药资讯,请关注CPHI制药在线▼点击阅读原文,进入智药研习社~
临床3期上市批准临床申请优先审批申请上市
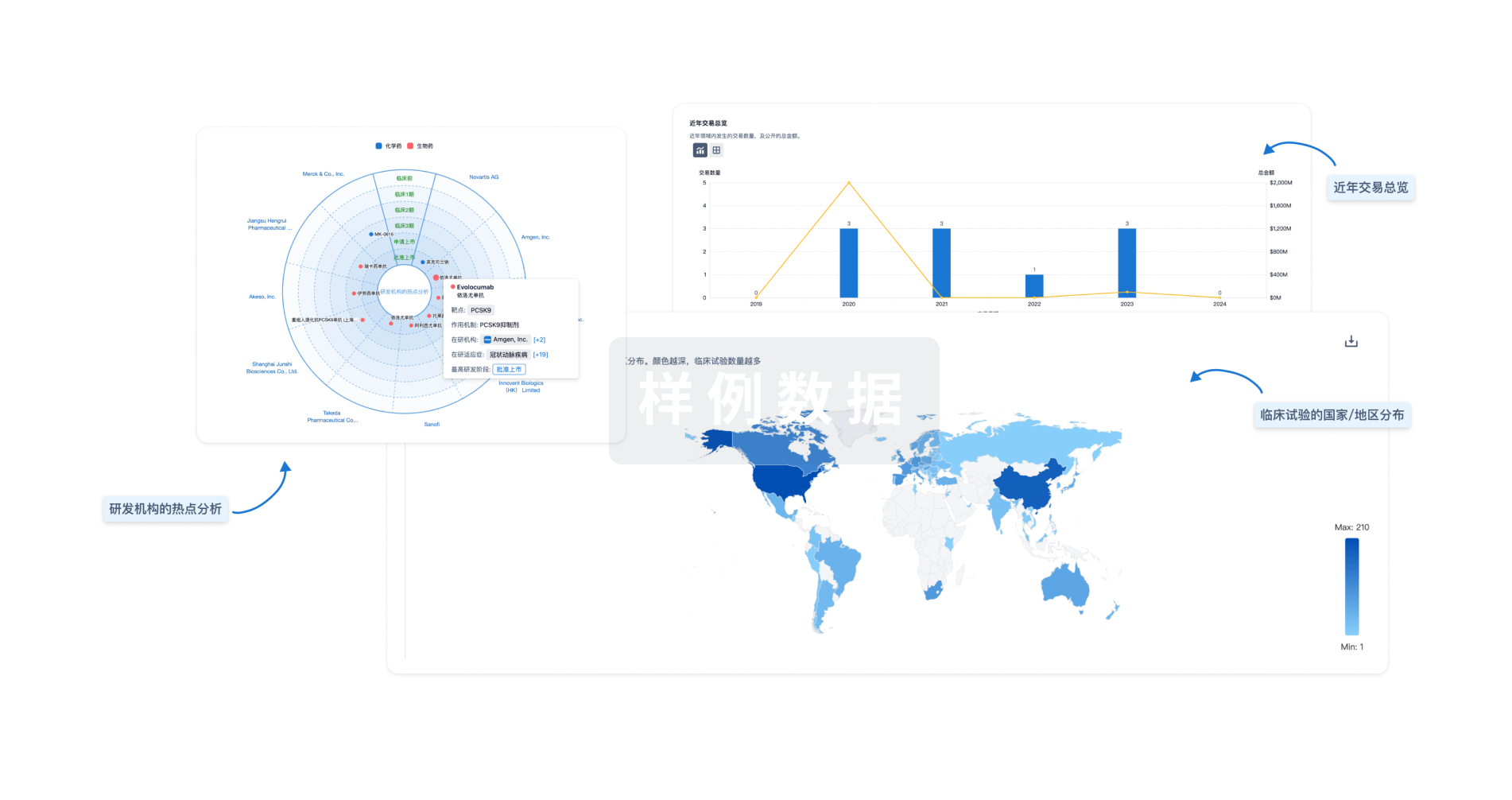
分析
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用



