预约演示
更新于:2025-05-07

Skaggs School of Pharmacy & Pharmaceutical Sciences
更新于:2025-05-07
概览
标签
消化系统疾病
肿瘤
皮肤和肌肉骨骼疾病
小分子化药
关联
1
项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的药物靶点 |
作用机制 CHD1L inhibitors |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
100 项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的临床结果
登录后查看更多信息
0 项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的专利(医药)
登录后查看更多信息
215
项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的文献(医药)2025-04-22·Journal of Chemical Theory and Computation
Computation of Protein–Ligand Binding Free Energies with a Quantum Mechanics-Based Mining Minima Algorithm
Article
作者: Guidez, Emilie B. ; Gordon, Mark S. ; Sattasathuchana, Tosaporn ; Webb, Simon P. ; Xu, Peng ; Potter, Michael J. ; Schlinsog, Megan ; Gilson, Michael K.
2025-04-08·Neurology
Acute Effects of Oral Levodopa on Seated Mean Arterial Blood Pressure in Parkinson’s Disease (P7-5.021)
作者: Litvan, Irene ; Moonla, Chochanon ; Longardner, Katherine ; Ghodsi, Hamid ; Momper, Jeremiah ; Wang, Joseph ; Mahato, Kuldeep ; Liu, Catherine
2025-04-08·Analytical Chemistry
Capillary Zone Electrophoresis-Mass Spectrometry of Intact G Protein-Coupled Receptors Enables Proteoform Profiling
Article
作者: Jooß, Kevin ; Kelleher, Neil L. ; Melani, Rafael D. ; Ives, Ashley N. ; Fellers, Ryan T. ; Janetzko, John
18
项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的新闻(医药)2024-12-03
TUESDAY, Dec. 3, 2024 -- Current prenatal multivitamin and mineral (PMVM) labels are misleading and often exclude the presence of heavy metals, according to a study published online Nov. 18 in the
American Journal of Clinical Nutrition
.
Laura M. Borgelt, Pharm.D., from the Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of Colorado Anschutz Medical Campus in Aurora, and colleagues analyzed 32 nonprescription and 15 prescription PMVMs regarding choline and iodine content, as well as arsenic, lead, and cadmium.
The researchers found that choline amounts were reported on 25.6 percent of products, including five within 20 percent of the claimed amount, two over the claimed amount by >20 percent, and five under the claimed amount by >20 percent. Iodine amounts were reported for 53.2 percent of products, including four within 20 percent of the claimed amount, 20 under the claimed amount by >20 percent, and one over the claimed amount by >20 percent. Amounts of arsenic, lead, and cadmium above United States Pharmacopeia purity limits were found in seven, two, and 13 PMVMs, respectively.
"Current PMVM labels are misleading with the potential to harm pregnant persons and fetuses through omission or inaccurate content of essential nutrients and inclusion of heavy metals," the authors write.
临床结果
2024-06-04
A group of researchers has identified the cause of a 'short-circuit' in cellular pathways, a discovery that sheds new light on the genesis of a number of human diseases and could lead to development of a wide array of new drugs.
A group of researchers at University of California San Diego has identified the cause of a "short-circuit" in cellular pathways, a discovery that sheds new light on the genesis of a number of human diseases.
The recent study, published in the journal Science Signaling, explores the biochemical mechanism that can interrupt the cellular communication chain -- a disruptive interaction that Pradipta Ghosh, M.D., likens to a game-ending "buzzer." Ghosh, a professor in the Departments of Medicine and Cellular and Molecular Medicine at University of California San Diego School of Medicine, and Irina Kufareva, Ph.D., an associate professor in the Skaggs School of Pharmacy and Pharmaceutical Sciences at University of California San Diego, are the corresponding authors on the paper.
The paper explains the mechanism of "cross talk" between two cellular pathways, one initiated by proteins known as growth factors and by their cellular receptors. The second pathway is mediated by a completely different G protein-coupled set of cellular receptors (GPCRs). Both classes of receptors deliver molecular messages from outside to inside the cell and signal cells to change in some way. Kufareva says that members of the GPCR family are targets of around 34 percent of all the drugs approved by the U.S. Food and Drug Administration.
"GPCRs are important drug targets mainly due to their involvement in signaling pathways related to many diseases," she explained, citing mental and endocrinological disorders, viral infections, cardiovascular and inflammatory conditions, and even cancer.
Growth factors enable a second, equally important communication pathway inside the cell that makes the cells grow and divide. Whereas GPCRs act through intracellular molecular switches (G proteins), growth factor receptors are conventionally thought to bypass the switches. However, Ghosh and Kufareva note that researchers had been suspicious about some kind of a potential conflict between the two pathways, and careful research allowed the UC San Diego team to identify it.
Ghosh said the conflict stems from problematic phosphorylation, the attachment of a phosphate group to the G protein molecule. She explained that the team used advanced mass spectrometry techniques to map all occurrences of phosphoevents, the sites on G proteins that were phosphorylated when cells were stimulated by growth factors. Then they checked how this changed the ability of G proteins to perform their normal job downstream of GPCRs.
"Whatever aspect of GPCR signaling we looked at, it was negatively impacted by almost all phosphoevents on the 'switch' protein -- the G protein -- that would be introduced by growth factors," Kufareva said. "That was understandable when we looked at how these phosphoevents distorted the G protein structure. Growth factors effectively 'steal' G proteins from GPCRs and in this way paralyze their signaling."
Further testing of the phosphoevents showed that one single amino acid was responsible for the G protein theft. Ghosh said the amino acid known as tyrosine is located at position 320 within the G protein, which happens to be on the side of the G protein that makes contact with G protein-coupled receptors.
"This specific tyrosine was identified almost a decade ago as a special 'trigger point' for G protein-coupled receptors to relay their signals. We began to think about the importance of such a coincidence," Ghosh explained. "That's when a light bulb went off in our heads: If cell communication were a game, the tyrosine at position 320 on the G protein would be the buzzer. If the growth factors got to it first and phosphorylated that site, the G protein-coupled receptors simply had no shot!"
Kufareva and Ghosh say that the group's discovery has implications for the development of new therapies for a number of conditions, including cancer. Ghosh said that many pharmaceuticals on the market are effective in treating a wide range of diseases because the drugs target G protein-coupled receptors. But there remain a number of conditions without good drug therapies -- fibrosis, chronic inflammation and cancers -- because until now the interaction of these two pathways has not been understood.
"We believe our findings are likely to be both important and timely, and will contribute to other emerging studies mapping the landscape of these two major signaling pathways that control practically every process in our cells," Ghosh said.
"Our work is especially relevant in that growth factors, their receptors, and G-protein-coupled receptors appear to be highly co-expressed in many cancers," added Kufareva.
All authors on the paper are associated with UC San Diego. Suchismita Roy, Saptarshi Sinha and Ananta James Silas are members of the School of Medicine's Department of Cellular and Molecular Medicine, while Majid Ghassemian is a member of the Department of Chemistry and Biochemistry, Biomolecular and Proteomics Mass Spectrometry Facility.
This paper was supported by the National Institutes of Health (CA238042, AI141630, CA100768 and CA160911 to Pradipta Ghosh; R21 AI149369, R01 GM136202, R21 AI156662, and R01 AI161880 to Irina Kufareva). Saptarshi Sinha was supported through the American Association of Immunologists Intersect Fellowship Program for Computational Scientist and Immunobiologists.
AACR会议
2024-03-11
Researchers have uncovered thousands of previously unknown bile acids, a type of molecule used by our gut microbiome to communicate with the rest of the body.
Researchers from Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California San Diego have uncovered thousands of previously unknown bile acids, a type of molecule used by our gut microbiome to communicate with the rest of the body.
"Bile acids are a key component of the language of the gut microbiome, and finding this many new types radically expands our vocabulary for understanding what our gut microbes do and how they do it," said senior author Pieter Dorrestein, Ph.D., professor at Skaggs School of Pharmacy and Pharmaceutical Sciences and professor of pharmacology and pediatrics at UC San Diego School of Medicine. "It's like going from 'See Spot Run' to Shakespeare."
The results, as described by study co-author and bile acids expert Lee Hagey, Ph.D, are akin to a molecular Rosetta stone, providing previously unknown insight into the biochemical language microbes use to influence distant organ systems.
Bile acids originate in the liver, are stored in the gallbladder and ultimately released into the intestine, where they are deployed to aid in digestion following the consumption of a meal. The microbes in our gut metabolize the bile acids produced by the liver, changing them into a vast array of different molecules called secondary bile acids, which tend to be easier for the body to absorb. Until now, the rich diversity and range of functions of secondary bile acids have been underappreciated by scientists.
"When I started working in the lab, there were about a few hundred known bile acids," said study co-author Ipsita Mohanty, Ph.D., a postdoctoral researcher in the Dorrestein lab. "Now we've discovered thousands more, and we're also working toward realizing that these bile acids do so much more than just help with digestion."
In addition to aiding digestion, bile acids are also important signaling molecules that help regulate the immune system and serve important metabolic functions, such as controlling lipid and glucose metabolism. These molecules also help explain how microbes in the gut are able to influence distant organ systems.
"Because of their interaction with our microbiome, the influence of bile acids spreads far beyond the digestive system, and so could the diseases we treat with them -- the list of diseases related to bile acids is a mile long, and there are several FDA approvals for these kinds of acids as treatments," said co-author Helena Mannochio-Russo, Ph.D., also a postdoctoral researcher in the Dorrestein lab.
In order to discover these molecules, the researchers leveraged the unique resources of UC San Diego. Dorrestein is director of the Collaborative Microbial Metabolite Center (CMMC), a first-of-its-kind collaboration between UC San Diego and UC Riverside that seeks to gather and centralize information about the metabolites that microbes produce to help researchers learn more about their impact on human health and the environment.
"In other areas of biology like genomics, sharing data is common, but there hasn't been an infrastructure in place for microbial metabolomics researchers to share data until now," said Dorrestein "Ultimately these breakthroughs are the result of a convergence of collaboration and computing power, and we expect many more breakthroughs to come out of the CMMC."
Earlier this year, the team debuted a new tool that can instantly match microbes to the metabolites they produce. The present study is the first of potentially many studies to utilize the tool for specific types of molecules. The researchers next hope to explore the specific functions of their newly-discovered bile acids as well as use their approach on other types of biomolecules, such as lipids or other kinds of acids.
"We're rewriting the textbook of human metabolism," said Dorrestein. "If you'd have spoken to me a few years ago, I would have said we were decades away from solving this puzzle, but now, it could happen within five years. It's really a remarkable change in our capabilities, and we believe it's going to revolutionize the way we approach disease."
微生物疗法
100 项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的药物交易
登录后查看更多信息
100 项与 Skaggs School of Pharmacy & Pharmaceutical Sciences 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年07月20日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
临床前
1
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
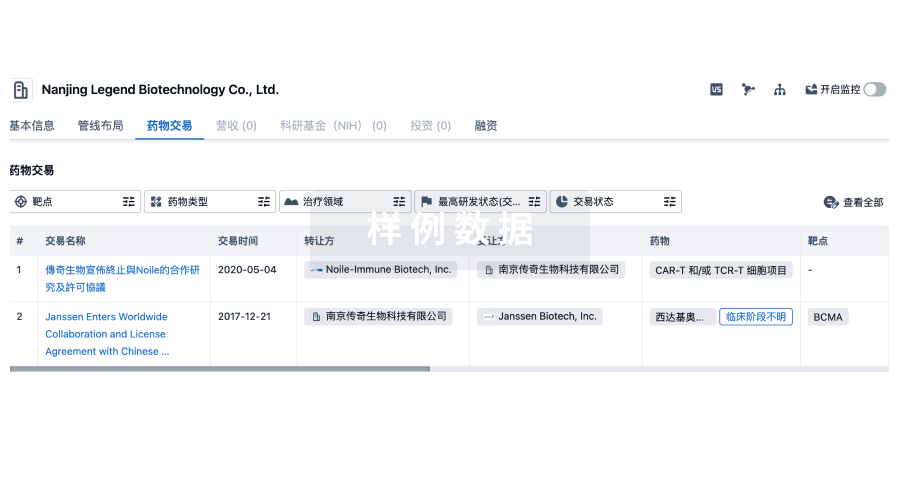
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
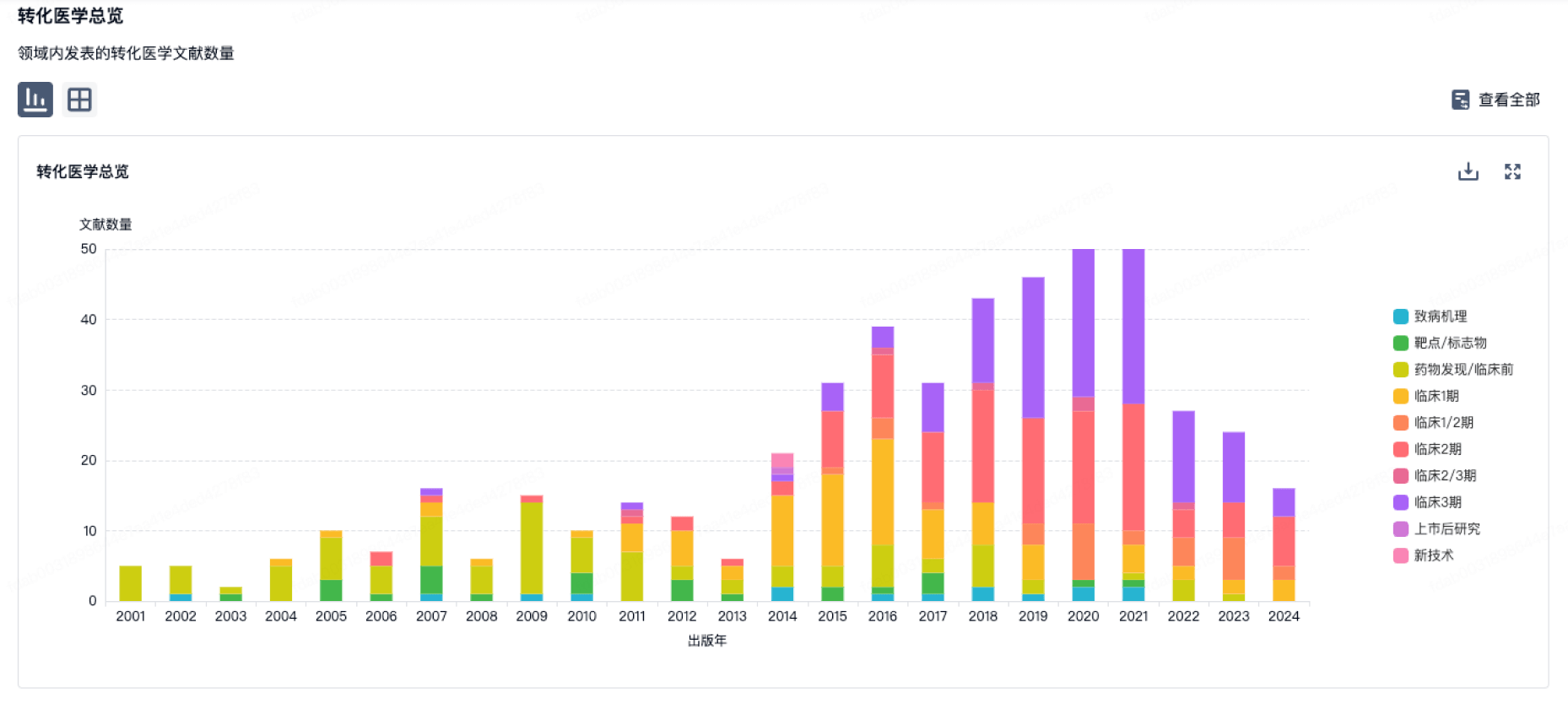
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
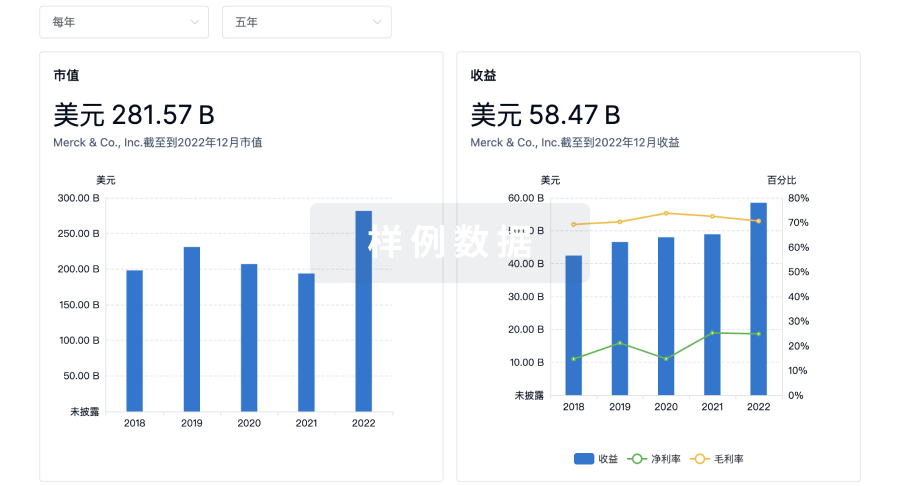
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用