预约演示
更新于:2025-05-07
Intervertebral Disc Degeneration
椎间盘退化
更新于:2025-05-07
基本信息
别名 Degenerated intervertebral disc、Degeneration of intervertebral disc、Degeneration of intervertebral disc (disorder) + [72] |
简介 Degenerative changes in the INTERVERTEBRAL DISC due to aging or structural damage, especially to the vertebral end-plates. |
关联
62
项与 椎间盘退化 相关的药物作用机制 硫酸软骨素蛋白聚糖调节剂 [+1] |
在研机构 |
原研机构 |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区 日本 |
首次获批日期2018-03-23 |
靶点 |
作用机制 TGF-β1调节剂 [+1] |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区 韩国 |
首次获批日期2017-07-01 |
1,172
项与 椎间盘退化 相关的临床试验NCT05971329
Pilot Study of ZetaFuse™ Bone Graft for the Treatment of Cervical Degenerative Disc Disease
The goal of this pilot clinical trial is to test the safety and preliminary performance of the ZetaFuse Bone Graft in patient requiring fusion of the C3-C7 vertebral bones due to pain or loss of neurological function.
Participants will be treated with ZetaFuse during surgical intervention to reduce pain and the loss of neurological function.
Participants will be treated with ZetaFuse during surgical intervention to reduce pain and the loss of neurological function.
开始日期2026-09-01 |
申办/合作机构 |
DRKS00031819
Sponsor-initiated, prospective, monocentric, non-interventional clinical observational study (AWB) to evaluate the Transmaxx® ALIF Cage in spine surgery use according to the intended purpose. - PMCF Transmaxx system
开始日期2025-12-31 |
申办/合作机构- |
NCT06144970
A Phase I, Double-blind, Randomized, Sham-controlled, Dose-response, Study Evaluating the Safety and Tolerability of TG-C in Subjects with Chronic Discogenic Lumbar Back Pain Due to Degenerative Disc Disease At 6 and 12 Months
The goal of this study is to evaluate the safety and tolerability of TG-C in subjects with chronic discogenic lumbar back pain due to degenerative disc disease. Participants will be administered a single intradiscal injection or subcutaneous injection for sham and followed up with in-clinic visits and telephone calls for 24 months.
开始日期2025-11-01 |
申办/合作机构 |
100 项与 椎间盘退化 相关的临床结果
登录后查看更多信息
100 项与 椎间盘退化 相关的转化医学
登录后查看更多信息
0 项与 椎间盘退化 相关的专利(医药)
登录后查看更多信息
17,926
项与 椎间盘退化 相关的文献(医药)2025-12-31·Veterinary Quarterly
High-intensity zones in dogs with lumbosacral intervertebral disc degeneration: insights from MRI and histopathological findings
Article
作者: van den Broek, Dirk Hendrik Nicolaas ; Tryfonidou, Marianna A. ; Kamali, S. Amir ; Meij, Björn P. ; Burgers, Elisabeth M. ; Teunissen, Michelle ; Grinwis, Guy C. M. ; Ito, Keita
2025-12-31·Annals of Medicine
A diagnostic signatures for intervertebral disc degeneration using TNFAIP6 and COL6A2 based on single-cell RNA-seq and bulk RNA-seq analyses
Article
作者: Shen, Jun-Kang ; Li, Zuo-Wei ; Li, Ming-Hua ; Song, Hong-Mei ; Wu, Chun-Gen ; Huang, Qin
2025-12-31·Annals of Medicine
Finite element models of intervertebral disc: recent advances and prospects
Review
作者: Sun, Tianze ; Wang, Jinzuo ; Wang, Junlin ; Liu, Xin ; Suo, Moran ; Li, Zhonghai ; Huang, Huagui ; Zhang, Jing
306
项与 椎间盘退化 相关的新闻(医药)2025-04-30
NEW YORK, April 29, 2025 (GLOBE NEWSWIRE) -- Mesoblast Limited (Nasdaq:MESO; ASX:MSB), global leader in allogeneic cellular medicines for inflammatory diseases, today provided highlights of its recent activities for the third quarter ended March 31, 2025. “We were very pleased to have made Ryoncil® (remestemcel-L) commercially available to treat children with acute GVHD within one quarter of receiving FDA approval as the first mesenchymal stromal cell (MSC) therapy approved in the US for any indication,” said Dr. Silviu Itescu, CEO of Mesoblast. “With our strong cash position we are well placed to expand Ryoncil® indications to other serious and life-threatening pediatric inflammatory diseases, and to adults with acute GvHD.” FINANCIAL HIGHLIGHTS Net operating cash spend for the quarter was US$12.7 million.Cash on hand at the end of the quarter was US$182 million (A$290 million)1. OPERATIONAL HIGHLIGHTS Ryoncil® (remestemcel-L) U.S. Launch for Steroid-Refractory Acute Graft Versus Host Disease Ryoncil® became commercially available for purchase in the United States on March 28, 2025, with Federal Medicaid coverage, and to date 15 infusion kits have been purchased for patients to start or continue their treatment course.Ryoncil® infusion kits are purchased and distributed by Cencora to enable the efficient and secure delivery of cryopreserved product to U.S. treatment centers, either directly or via a specialty pharmacy option.To date, ten priority transplant centers have been fully onboarded, five of whom have enrolled patients through the MyMesoblast™ hub.Mesoblast anticipates onboarding an additional ten priority transplant centers in the current quarter.The full team of nine key account managers (KAMs) commenced activities in the last week of April. The KAMs will accelerate onboarding of the remaining 35 priority transplant centers, accounting for 80% of U.S. pediatric transplants, and will drive the business to provide on the ground engagement with healthcare providers and administrators.Mesoblast has continued to expand coverage for Ryoncil® to over 104 million US lives insured by commercial and government payers.To date, 37 of the 51 States provide fee-for-service Medicaid coverage for Ryoncil® through Orphan Drug Lists or medical exception / prior authorization (PA) process. The remainder will come online July 1, 2025, with mandatory coverage for all 44 million lives.To assist patients and institutions with insurance coverage, financial assistance, and access programs, ensuring that no patient is left behind in receiving this potentially life-saving therapy, Mesoblast has established a patient access hub termed MyMesoblast™, where Ryoncil® is now available for ordering. Additional information is available on ryoncil.com, where valuable resources for healthcare providers, patients and caregivers can be found. Revascor® (rexlemestrocel-L) for Chronic Heart Failure with Reduced Ejection Fraction (HFrEF) and Persistent Inflammation Mesoblast has a Type B meeting with FDA scheduled for this quarter to discuss the accelerated approval pathway for Revascor® (rexlemestrocel-L) in the treatment of patients with ischemic chronic heart failure with reduced ejection fraction (HFrEF) and inflammation. The meeting will be held under Mesoblast’s Regenerative Medicines Advanced Therapy (RMAT) designation for REVASCOR.In a Type B meeting last year, FDA provided guidance to Mesoblast that the company was eligible to file for accelerated approval of REVASCOR in patients with end-stage HFrEF based on the totality of data across two randomized controlled trials. FDA also guided that a single confirmatory trial in class II/III patients with ischemic HFrEF and inflammation will need to be completed after any accelerated approval is obtained.The key objectives of the meeting are to obtain FDA feedback on relevant chemistry, manufacturing & controls (CMC), alignment on potency assays for commercial product release, and Mesoblast’s proposed design and primary endpoint for the confirmatory trial.In November 2024 a publication in the prestigious peer-reviewed European Journal of Heart Failure (EJHF) reported that a single intramyocardial injection of REVASCOR results in improved survival in high-risk NYHA Class II/III patients with ischemic heart failure and inflammation.2 This identifies the HFrEF population that is responsive to REVASCOR and will be the target of a confirmatory trial after accelerated approval, if received. Rexlemestrocel-L for Chronic Low Back Pain associated with Degenerative Disc Disease – Phase 3 Program The confirmatory Phase 3 trial of Mesoblast’s second generation allogeneic, STRO3-immunoselected, and industrially manufactured stromal cell product candidate rexlemestrocel-L in patients with chronic low back pain (CLBP) due to inflammatory degenerative disc disease (DDD) of less than five years duration is actively enrolling and treating patients at multiple sites across the U.S.FDA has previously agreed on the design of this 300-patient randomized, placebo-controlled confirmatory Phase 3 trial, and the 12-month primary endpoint of pain reduction as an approvable indication.This endpoint was successfully met in Mesoblast’s first Phase 3 trial. Key secondary measures include improvement in quality of life and function.A particular focus is on treatment of patients on opioids, since discogenic back pain accounts for approximately 50% of prescription opioid usage in the US. Significant pain reduction and opioid cessation were observed in Mesoblast’s first Phase 3 trial.FDA has designated rexlemestrocel-L a RMAT for the treatment of chronic low back pain. Corporate During the period, Mesoblast successfully completed a global private placement primarily to existing major US, UK, and Australian shareholders raising A$260 million (US$161 million).Strengthened Board of Directors with appointment of Dr. Gregory George and Ms Lyn Cobley.Mesoblast was added to the S&P Dow Jones Indices’ S&P/ASX 200 Index effective March 6, 2025, on the Australian Stock Exchange (ASX). Other Fees to Non-Executive Directors were US$50,306, consulting payments to Non-Executive Directors were Nil and salary payments to full-time Executive Directors were US$223,092, detailed in Item 6 of the Appendix 4C cash flow report for the quarter.3 From August 2023 to July 2025, our Non-Executive Directors have voluntarily reduced cash payment of their fees by 50% and Executive Directors (our Chief Executive and Chief Medical Officers) reduced their base salaries by 30%, in lieu of accepting equity-based incentives. A copy of the Appendix 4C – Quarterly Cash Flow Report for the third quarter FY2025 is available on the investor page of the company’s website www.mesoblast.com. About Mesoblast Mesoblast (the Company) is a world leader in developing allogeneic (off-the-shelf) cellular medicines for the treatment of severe and life-threatening inflammatory conditions. The therapies from the Company’s proprietary mesenchymal lineage cell therapy technology platform respond to severe inflammation by releasing anti-inflammatory factors that counter and modulate multiple effector arms of the immune system, resulting in significant reduction of the damaging inflammatory process. Mesoblast’s RYONCIL® (remestemcel-L) for the treatment of steroid-refractory acute graft versus host disease (SR-aGvHD) in children 2 months and older is the first FDA-approved mesenchymal stromal cell (MSC) therapy. Please see the full Prescribing Information at www.ryoncil.com. Mesoblast is committed to developing additional cell therapies for distinct indications based on its remestemcel-L and rexlemestrocel-L allogeneic stromal cell technology platforms. RYONCIL is being developed for additional inflammatory diseases including SR-aGvHD in adults and biologic-resistant inflammatory bowel disease. Rexlemestrocel-L is being developed for heart failure and chronic low back pain. The Company has established commercial partnerships in Japan, Europe and China. About Mesoblast intellectual property: Mesoblast has a strong and extensive global intellectual property portfolio, with over 1,000 granted patents or patent applications covering mesenchymal stromal cell compositions of matter, methods of manufacturing and indications. These granted patents and patent applications provide commercial protection extending through to at least 2041 in all major markets. About Mesoblast manufacturing: The Company’s proprietary manufacturing processes yield industrial-scale, cryopreserved, off-the-shelf, cellular medicines. These cell therapies, with defined pharmaceutical release criteria, are planned to be readily available to patients worldwide. Mesoblast has locations in Australia, the United States and Singapore and is listed on the Australian Securities Exchange (MSB) and on the Nasdaq (MESO). For more information, please see www.mesoblast.com, LinkedIn: Mesoblast Limited and Twitter: @Mesoblast References / Footnotes Translated at 1A$:0.6413US$ being the March 31, 2025 rate as reported by the Reserve Bank of Australia.Perin EC. Et al. Mesenchymal precursor cells reduce mortality and major morbidity in ischaemic heart failure with inflammation: DREAM-HF. Eur J Heart Fail 2024. https://doi-org.libproxy1.nus.edu.sg/10.1002/ejhf.3522As required by ASX listing rule 4.7 and reported in Item 6 of the Appendix 4C, reported are the aggregated total payments to related parties being Executive Directors and Non-Executive Directors. Forward-Looking StatementsThis press release includes forward-looking statements that relate to future events or our future financial performance and involve known and unknown risks, uncertainties and other factors that may cause our actual results, levels of activity, performance or achievements to differ materially from any future results, levels of activity, performance or achievements expressed or implied by these forward-looking statements. We make such forward-looking statements pursuant to the safe harbor provisions of the Private Securities Litigation Reform Act of 1995 and other federal securities laws. Forward-looking statements should not be read as a guarantee of future performance or results, and actual results may differ from the results anticipated in these forward-looking statements, and the differences may be material and adverse. Forward-looking statements include, but are not limited to, statements about: the initiation, timing, progress and results of Mesoblast’s preclinical and clinical studies, and Mesoblast’s research and development programs; Mesoblast’s ability to advance product candidates into, enroll and successfully complete, clinical studies, including multi-national clinical trials; Mesoblast’s ability to advance its manufacturing capabilities; the timing or likelihood of regulatory filings and approvals, manufacturing activities and product marketing activities, if any; the commercialization of Mesoblast’s RYONCIL for pediatric SR-aGVHD and any other product candidates, if approved; regulatory or public perceptions and market acceptance surrounding the use of stem-cell based therapies; the potential for Mesoblast’s product candidates, if any are approved, to be withdrawn from the market due to patient adverse events or deaths; the potential benefits of strategic collaboration agreements and Mesoblast’s ability to enter into and maintain established strategic collaborations; Mesoblast’s ability to establish and maintain intellectual property on its product candidates and Mesoblast’s ability to successfully defend these in cases of alleged infringement; the scope of protection Mesoblast is able to establish and maintain for intellectual property rights covering its product candidates and technology; estimates of Mesoblast’s expenses, future revenues, capital requirements and its needs for additional financing; Mesoblast’s financial performance; developments relating to Mesoblast’s competitors and industry; and the pricing and reimbursement of Mesoblast’s product candidates, if approved. You should read this press release together with our risk factors, in our most recently filed reports with the SEC or on our website. Uncertainties and risks that may cause Mesoblast’s actual results, performance or achievements to be materially different from those which may be expressed or implied by such statements, and accordingly, you should not place undue reliance on these forward-looking statements. We do not undertake any obligations to publicly update or revise any forward-looking statements, whether as a result of new information, future developments or otherwise. Release authorized by the Chief Executive. For more information, please contact: Corporate Communications / InvestorsPaul HughesT: +61 3 9639 6036 Media – Global Allison WorldwideEmma NealT: +1 603 545 4843E: emma.neal@allisonworldwide.com Media – AustraliaBlueDot MediaSteve DabkowskiT: +61 419 880 486E: steve@bluedot.net.au
临床3期细胞疗法加速审批财报高管变更
2025-04-29
HOUSTON, April 29, 2025 (GLOBE NEWSWIRE) -- FibroBiologics, Inc. (Nasdaq: FBLG) (“FibroBiologics”), a clinical-stage biotechnology company with 240+ patents issued and pending with a focus on the development of therapeutics and potential cures for chronic diseases using fibroblasts and fibroblast-derived materials, today announced that Simon Gebremeskel, research scientist at FibroBiologics, is presenting a poster at the ThymUS 2025 Meeting taking place in Lihue, Kaua'i Hawaii from April 27 - May 1, 2025. Founded in 2001, ThymUS brings together international researchers working on thymus and T-cell biology. The abstract entitled “A rapid and scalable thymus organoid development method for generating functional T cells” presents progress on the development of a transplantable and scalable artificial thymus organoid. The poster can be viewed on FibroBiologics’ website here. “Our latest research demonstrates the potential of using an artificial thymus organoid to recover the lost functionality of the thymus, an organ that is the T-cell teaching center of the immune system,” said Hamid Khoja, Ph.D., Chief Scientific Officer of FibroBiologics. “Organoids are transforming how we study and develop potential treatments for multiple chronic diseases. The unique immunomodulatory properties of fibroblasts offer promising potential in ensuring the functionality and durability of therapeutic organoids.” For more information, please visit FibroBiologics' website or email FibroBiologics at: info@fibrobiologics.com. Cautionary Statement Regarding Forward-Looking Statements This communication contains "forward-looking statements" as defined in the Private Securities Litigation Reform Act of 1995. Forward-looking statements include information concerning the potential and capabilities of fibroblasts and artificial thymus organoids to recover the lost functionality of the thymus. These forward-looking statements are based on FibroBiologics' management's current expectations, estimates, projections and beliefs, as well as a number of assumptions concerning future events. When used in this communication, the words "estimates," "projected," "expects," "anticipates," "forecasts," "plans," "intends," "believes," "seeks," "may," "will," "should," "future," "propose" and variations of these words or similar expressions (or the negative versions of such words or expressions) are intended to identify forward-looking statements. These forward-looking statements are not guarantees of future performance, conditions or results, and involve a number of known and unknown risks, uncertainties, assumptions and other important factors, many of which are outside FibroBiologics' management's control, that could cause actual results to differ materially from the results discussed in the forward-looking statements, including those set forth under the caption "Risk Factors" and elsewhere in FibroBiologics' annual, quarterly and current reports (i.e., Form 10-K, Form 10-Q and Form 8-K) as filed or furnished with the SEC and any subsequent public filings. Copies are available on the SEC's website, www.sec.gov. These risks, uncertainties, assumptions and other important factors include, but are not limited to: (a) expectations regarding the initiation, progress and expected results of our R&D efforts and preclinical studies; (b) the unpredictable relationship between R&D and preclinical results and clinical study results; (c) risks related to FibroBiologics' liquidity and its ability to maintain capital resources sufficient to conduct its business, and (d) the ability of FibroBiologics to successfully prosecute its patent applications. Forward-looking statements speak only as of the date they are made. Readers are cautioned not to put undue reliance on forward-looking statements, and FibroBiologics assumes no obligation and, except as required by law, does not intend to update or revise these forward-looking statements, whether as a result of new information, future events, or otherwise. FibroBiologics gives no assurance that it will achieve its expectations. About FibroBiologics Based in Houston, FibroBiologics is a clinical-stage biotechnology company developing a pipeline of treatments and seeking potential cures for chronic diseases using fibroblast cells and fibroblast-derived materials. FibroBiologics holds 240+ US and internationally issued patents/patents pending across various clinical pathways, including wound healing, multiple sclerosis, disc degeneration, psoriasis, orthopedics, human longevity, and cancer. FibroBiologics represents the next generation of medical advancement in cell therapy and tissue regeneration. For more information, visit www.FibroBiologics.com. General Inquiries: info@fibrobiologics.com Investor Relations: Nic Johnson Russo Partners (212) 845-4242 fibrobiologicsIR@russopr.com Media Contact: Liz Phillips Russo Partners (347) 956-7697 Elizabeth.phillips@russopartnersllc.com
细胞疗法
2025-04-11
HOUSTON, April 11, 2025 (GLOBE NEWSWIRE) -- FibroBiologics, Inc. (Nasdaq: FBLG) ("FibroBiologics”), a clinical-stage biotechnology company with 240+ patents issued and pending for the development of therapeutics and potential cures for chronic diseases using fibroblasts and fibroblast-derived materials, today announced that Hamid Khoja, Ph.D., Chief Scientific Officer of FibroBiologics, will present on FibroBiologics’ unique fibroblast cell-based approach for the potential treatment of chronic diseases at The Cell & Gene Meeting On The Mediterranean (Med) in Rome, Italy, from April 15-17, 2025. The Cell & Gene Meeting on the Med is the leading conference bringing together the Advanced Therapy Medicinal Products community from Europe and beyond. Details of the conference and presentations are as follows: The Cell & Gene Meeting On The Med
Presentation Title: Restoring Immune system homeostasis using a fibroblast cell-based therapeutic targeting three chronic inflammation-mediated diseases Presenter: Hamid Khoja, Ph.D., Chief Scientific Officer, FibroBiologics Location: Salone dei Cavalieri, Section 2 Date and Time: Wednesday, April 16 at 9:00 AM CEST For more information, please visit FibroBiologics' website or email FibroBiologics at: info@fibrobiologics.com. About FibroBiologics Based in Houston, FibroBiologics is a clinical-stage biotechnology company developing a pipeline of treatments and potential cures for chronic diseases using fibroblast cells and fibroblast-derived materials. FibroBiologics holds 240+ US and internationally issued patents/patents pending across various clinical pathways, including wound healing, multiple sclerosis, disc degeneration, psoriasis, orthopedics, human longevity, and cancer. FibroBiologics represents the next generation of medical advancement in cell therapy and tissue regeneration. For more information, visit www.FibroBiologics.com. General Inquiries:info@fibrobiologics.com Investor Relations:Nic JohnsonRusso Partners(212) 845-4242fibrobiologicsIR@russopr.com Media Contact:Liz PhillipsRusso Partners(347) 956-7697Elizabeth.phillips@russopartnersllc.com
基因疗法
分析
对领域进行一次全面的分析。
登录
或
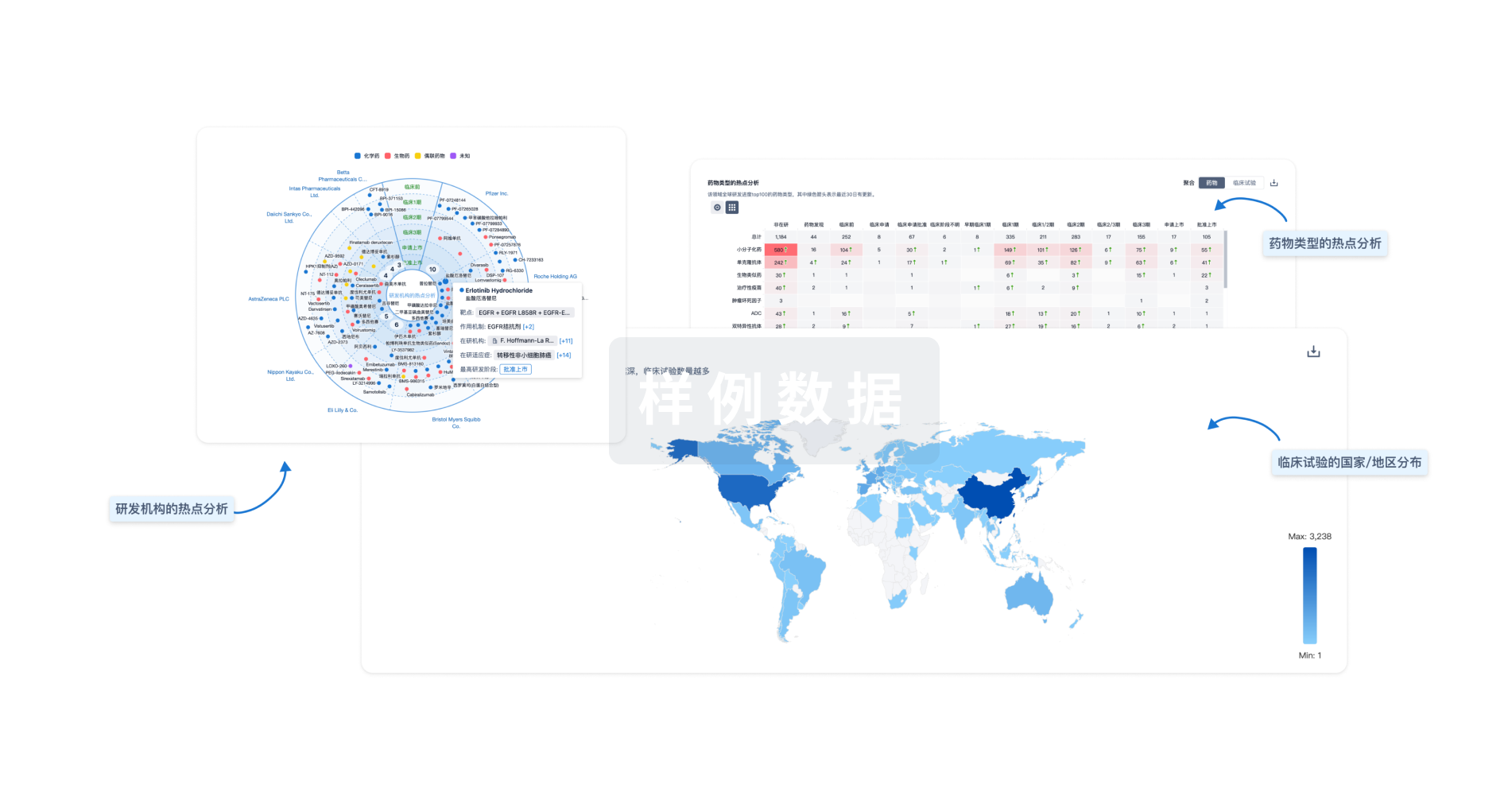
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用



