预约演示
更新于:2025-05-07
Bone Marrow Neoplasms
骨髓肿瘤
更新于:2025-05-07
基本信息
别名 Bone Marrow Neoplasm、Bone Marrow Neoplasms、Bone Marrow Tumor + [16] |
简介 Neoplasms located in the bone marrow. They are differentiated from neoplasms composed of bone marrow cells, such as MULTIPLE MYELOMA. Most bone marrow neoplasms are metastatic. |
关联
154
项与 骨髓肿瘤 相关的药物靶点 |
作用机制 CSF-1R拮抗剂 |
在研机构 |
原研机构 |
非在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2024-08-14 |
作用机制 端粒末端转移酶抑制剂 |
在研机构 |
原研机构 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2024-06-06 |
靶点 |
作用机制 HDAC抑制剂 |
在研机构 |
原研机构 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2024-03-21 |
530
项与 骨髓肿瘤 相关的临床试验NCT06930651
A Phase I/II Study of CAR.70-Engineered IL15-Transduced Cord Blood-Derived NK Cells With TGF-beta Receptor 2 (TGFBR2) Knock Out in Conjunction With Lymphodepleting Chemotherapy for the Management of Relapsed/Refractory Myeloid Malignancies
The goal of this clinical research study is to find the recommended safe dose of TGFBR2 KO CAR27/IL-15 NK cells that can be given to patients with relapsed/refractory disease. The safety and effectiveness of this treatment will also be studied.
开始日期2025-10-01 |
NCT06928662
Sequential Decitabine in Combination With FLAG-Ida Followed Immediately by Reduced-Intensity Conditioning (RIC) Allogeneic Hematopoietic Cell Transplantation (DEC-FLAG-Ida/RIC) for Adults With Myeloid Malignancies at High Risk of Relapse: A Phase 1/2 Study
This phase I/II trial studies the safety, side effects, and best dose of decitabine in combination with fludarabine, cytarabine, filgrastim, and idarubicin (FLAG-Ida) and total body irradiation (TBI) followed by a donor stem cell transplant in treating adult patients with cancers of blood-forming cells of the bone marrow (myeloid malignancies) that are at high risk of coming back after treatment (relapse). Cancers eligible for this trial are acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and chronic myelomonocytic leukemia (CMML). Decitabine is in a class of medications called hypomethylation agents. It works by helping the bone marrow produce normal blood cells and by killing abnormal cells in the bone marrow. The FLAG-Ida regimen consists of the following drugs: fludarabine, cytarabine, filgrastim, and idarubicin. These are chemotherapy drugs that work in different ways to stop the growth of cancer cells, either by killing the cells, by stopping them from dividing, or by stopping them from spreading. Filgrastim is in a class of medications called colony-stimulating factors. It works by helping the body make more neutrophils, a type of white blood cell. Radiation therapy uses high energy x-rays, particles, or radioactive seeds to kill cancer cells and shrink tumors. TBI is radiation therapy to the entire body. Giving chemotherapy and TBI before a donor peripheral blood stem cell (PBSC) transplant helps kill cancer cells in the body and helps make room in the patient's bone marrow for new blood-forming cells (stem cells) to grow. When the healthy stem cells from a donor are infused into a patient, they may help the patient's bone marrow make more healthy cells and platelets. Giving decitabine in combination with FLAG-Ida and TBI before donor PBSC transplant may work better than FLAG-Ida and TBI alone in treating adult patients with myeloid malignancies at high risk of relapse.
开始日期2025-09-01 |
NCT06710418
Red Blood Cell Transfusion Threshold-Specific Bleeding, Quality of Life and Functional Outcomes in Acute Leukemia Patients With Thrombocytopenia: a Randomized Feasibility Study
This clinical trial evaluates the effects of hemoglobin threshold-specific packed red blood cell (PRBC) transfusions on quality of life and functional outcomes in patients who have undergone chemotherapy or an allogeneic hematopoietic stem cell transplant for a high-grade myeloid neoplasm, acute myeloid leukemia, or B acute lymphoblastic lymphoma/leukemia. Some types of chemotherapy and stem cell transplants can induce low platelet counts and/or anemia that requires PRBC transfusions. Given critical shortages in blood supply, and risks associated with transfusion of PRBC, there has been much investigation into the "minimum" hemoglobin level that effectively balances safety and toxicity in patients. This clinical trial evaluates the effects of giving PRBC transfusions based on a more restrictive hemoglobin threshold (> 7 gm/dL) compared to a more liberal hemoglobin threshold (> 9 gm/dL) on quality of life and functional outcomes. A more restrictive threshold may be just as effective at maintaining patient quality of life and function while decreasing side effects from blood transfusions and helping to conserve blood supply resources.
开始日期2025-07-01 |
申办/合作机构 |
100 项与 骨髓肿瘤 相关的临床结果
登录后查看更多信息
100 项与 骨髓肿瘤 相关的转化医学
登录后查看更多信息
0 项与 骨髓肿瘤 相关的专利(医药)
登录后查看更多信息
16,695
项与 骨髓肿瘤 相关的文献(医药)2025-12-31·mAbs
T cell margination: investigating the detour of T cells following forimtamig treatment in humanized mice
Article
作者: O’Brien, Nils ; Lechner, Stefanie ; Eckmann, Jan ; Osl, Franz ; Klein, Christian ; Pöschinger, Thomas ; Attig, Jan ; Wolf, Ann-Katrin ; Mueller, Joerg P.J. ; Bröske, Ann-Marie E. ; Colombetti, Sara ; Rae, Richard ; Beilhack, Andreas ; Umaña, Pablo ; Crisand, Cylia
2025-12-31·Hematology
Identification of t(X;1)(q28;q21) generating a novel GATAD2B::MTCP1 gene fusion in CMML and its persistence during progression to AML
Article
作者: Chen, Su-ning ; Chen, Yu ; Liu, Yi-zi ; Zhang, Zhi-yu ; Hou, Chun-xiao ; Zhang, Feng-hong ; Wang, Qian ; Zhu, Yi-yan
2025-12-31·Hematology
RUNX1::MECOM
rearrangement in myeloid neoplasm post cytotoxic therapy following sarcoma treatment: a case presentation and review of the literature
Review
作者: Brendsdal Forthun, Rakel ; Dahl Hamnvik, Lars Henrik ; Kollsete Gjelberg, Hilde ; Sefland, Øystein ; Andersson Tvedt, Tor Henrik ; Sandnes, Miriam ; Reikvam, Håkon ; Ktoridou-Valen, Irini
1,094
项与 骨髓肿瘤 相关的新闻(医药)2025-05-01
– U.S. Regulatory Filing Under Active Review for Approval of PYRUKYND® (mitapivat) in Thalassemia, with PDUFA Goal Date of September 7, 2025 –
– Phase 3 RISE UP Study of Mitapivat in Sickle Cell Disease On Track, with Topline Results Expected in Late 2025; Potential U.S. Commercial Launch in 2026 –
– Tebapivat Advancing in Clinical Trials for Lower-Risk Myelodysplastic Syndromes (LR-MDS) and Sickle Cell Disease –
– PYRUKYND Net Revenue of $8.7 Million in Q1; Cash, Cash Equivalents and Marketable Securities of $1.4 Billion as of March 31, 2025 –
CAMBRIDGE, Mass. , May 01, 2025 (GLOBE NEWSWIRE) -- Agios Pharmaceuticals, Inc. (Nasdaq: AGIO), a leader in cellular metabolism and pyruvate kinase (PK) activation pioneering therapies for rare diseases, today reported business highlights and financial results for the first quarter ended March 31, 2025 .
“We are pleased with our strong start to 2025, highlighted by the acceptance of our sNDA for thalassemia with a PDUFA goal date of September 7, 2025 . Our engagement with the FDA is progressing as expected, and we are committed to bringing PYRUKYND to thalassemia patients, irrespective of genotype or transfusion needs,” said Brian Goff , chief executive officer at Agios. “Looking ahead, our focus is also on delivering the topline results from the Phase 3 RISE UP study in sickle cell disease, which remains on track for year-end, and continuing to advance our early and mid-stage clinical programs. Supported by our strong financial position and highly experienced team, we are driving forward PYRUKYND’s multi-billion-dollar potential while building a pipeline designed for lasting impact, with the goal of creating significant value for shareholders and delivering transformative therapies for patients.”
First Quarter 2025 and Recent Highlights
PYRUKYND® Revenues: Generated $8.7 million in net revenue for the first quarter of 2025, compared to $8.2 million in the first quarter of 2024. A total of 234 unique patients have completed prescription enrollment forms, representing an increase of 5 percent over the fourth quarter of 2024. A total of 136 patients are on PYRUKYND therapy, inclusive of new prescriptions and continued therapy, as compared to 130 patients at the end of the fourth quarter 2024. Thalassemia: The U.S. Food and Drug Administration (FDA) accepted the company’s supplemental New Drug Application (sNDA) for PYRUKYND for the treatment of adult patients with non-transfusion-dependent and transfusion-dependent alpha- or beta-thalassemia. The Prescription Drug User Fee Act (PDUFA) goal date is September 7, 2025. The FDA has communicated that at this time no advisory committee meeting is planned, and the review is ongoing. Sickle Cell Disease: The Phase 3 RISE UP study evaluating mitapivat for the treatment of sickle cell disease patients who are 16 years of age or older continued to progress as anticipated. This 52-week Phase 3 study completed enrollment in October 2024 , enrolling more than 200 patients worldwide. Advanced final preparations to initiate a Phase 2 clinical trial of tebapivat in patients with sickle cell disease in mid-2025. Pediatric Pyruvate Kinase (PK) Deficiency: Reported positive topline results from the ACTIVATE-Kids Phase 3 study of mitapivat in children aged 1 to <18 years with PK deficiency who are not regularly transfused. Safety was consistent with the profile for mitapivat previously observed in adults with PK deficiency who are not regularly transfused. ACTIVATE-Kids is the first study to demonstrate efficacy of an oral therapy for children with PK Deficiency who are not regularly transfused. Lower-risk Myelodysplastic Syndromes (LR-MDS): Progressed patient enrollment in the Phase 2b study of tebapivat in LR-MDS. Corporate: Krishnan Viswanadhan , Pharm. D, joined Agios as Chief Corporate Development and Strategy Officer, responsible for leading the company’s corporate strategy, business development, and long-term growth initiatives. Previously, he served as President and Chief Operating Officer of Be Biopharma and at various senior roles at both Bristol Myers Squibb and Celgene.
The U.S. Food and Drug Administration (FDA) accepted the company’s supplemental New Drug Application (sNDA) for PYRUKYND for the treatment of adult patients with non-transfusion-dependent and transfusion-dependent alpha- or beta-thalassemia. The Prescription Drug User Fee Act (PDUFA) goal date is September 7, 2025. The FDA has communicated that at this time no advisory committee meeting is planned, and the review is ongoing.
The Phase 3 RISE UP study evaluating mitapivat for the treatment of sickle cell disease patients who are 16 years of age or older continued to progress as anticipated. This 52-week Phase 3 study completed enrollment in October 2024 , enrolling more than 200 patients worldwide. Advanced final preparations to initiate a Phase 2 clinical trial of tebapivat in patients with sickle cell disease in mid-2025.
Reported positive topline results from the ACTIVATE-Kids Phase 3 study of mitapivat in children aged 1 to <18 years with PK deficiency who are not regularly transfused. Safety was consistent with the profile for mitapivat previously observed in adults with PK deficiency who are not regularly transfused. ACTIVATE-Kids is the first study to demonstrate efficacy of an oral therapy for children with PK Deficiency who are not regularly transfused.
Progressed patient enrollment in the Phase 2b study of tebapivat in LR-MDS.
Key Upcoming Milestones & Priorities
Agios expects to achieve the following key milestones in 2025:
Thalassemia: Receive FDA regulatory decision for PYRUKYND for the treatment of adult patients with non-transfusion-dependent and transfusion-dependent alpha- or beta-thalassemia (PDUFA goal date is September 7, 2025). Continue progressing the review of regulatory applications with health authorities in the European Union , Kingdom of Saudi Arabia and United Arab Emirates . Sickle Cell Disease: Announce topline results from the Phase 3 RISE UP study of mitapivat in sickle cell disease in late 2025, with a potential U.S. commercial launch in 2026. Additionally, begin patient enrollment for the Phase 2 study of tebapivat in sickle cell disease in mid-2025. LR-MDS: Complete patient enrollment in the Phase 2b study of tebapivat for LR-MDS in late 2025. Early-Stage Pipeline: File an Investigational New Drug Application for AG-236, an siRNA targeting TMPRSS6 intended for the treatment of polycythemia vera, in mid-2025.
First Quarter 2025 Financial Results
Revenue: Net product revenue from sales of PYRUKYND for the first quarter of 2025 was $8.7 million , compared to $8.2 million for the first quarter of 2024.
Cost of Sales: Cost of sales for the first quarter of 2025 was $1.1 million .
Research and Development (R&D) Expenses: R&D expenses were $72.7 million for the first quarter of 2025, compared to $68.6 million for the first quarter of 2024. The year-over-year increase was primarily attributed to an increase in workforce-related expenses and costs associated with clinical trials of tebapivat in LR-MDS and sickle cell disease, partially offset by lower costs associated with the clinical trials of mitapivat in thalassemia and pediatric PKD.
Selling, General and Administrative (SG&A) Expenses: SG&A expenses were $41.5 million for the first quarter of 2025 compared to $31.0 million for the first quarter of 2024. The year-over-year increase was primarily attributable to an increase in commercial-related activities, including headcount, as the company prepares for the potential approval of PYRUKYND in thalassemia.
Net Loss: Net loss was $89.3 million for the first quarter of 2025 compared to $81.5 million for the first quarter of 2024.
Cash Position and Guidance: Cash, cash equivalents and marketable securities as of March 31, 2025 , were $1.4 billion compared to $1.5 billion as of December 31, 2024 . Agios expects that its cash, cash equivalents and marketable securities, together with anticipated product revenue and interest income, will provide the financial independence to prepare for potential PYRUKYND launches in thalassemia and sickle cell disease, advance existing programs, and to opportunistically expand its pipeline through both internally and externally discovered assets.
Conference Call Information
Agios will host a conference call and live webcast today at 8:00 a.m. ET to discuss the company’s first quarter 2025 financial results and recent business highlights. The live webcast will be accessible on the Investors section of the company's website (www.agios.com) under the “Events & Presentations” tab. A replay of the webcast will be available on the company’s website approximately two hours after the event.
About Agios
Agios is the pioneering leader in PK activation and is dedicated to developing and delivering transformative therapies for patients living with rare diseases. In the U.S. , Agios markets a first-in-class pyruvate kinase (PK) activator for adults with PK deficiency, the first disease-modifying therapy for this rare, lifelong, debilitating hemolytic anemia. Building on the company's deep scientific expertise in classical hematology and leadership in the field of cellular metabolism and rare hematologic diseases, Agios is advancing a robust clinical pipeline of investigational medicines with programs in alpha- and beta-thalassemia, sickle cell disease, pediatric PK deficiency, myelodysplastic syndromes (MDS)-associated anemia and phenylketonuria (PKU). In addition to its clinical pipeline, Agios is advancing a preclinical TMPRSS6 siRNA as a potential treatment for polycythemia vera. For more information, please visit the company’s website at www.agios.com.
Cautionary Note Regarding Forward-Looking Statements
This press release contains forward-looking statements within the meaning of The Private Securities Litigation Reform Act of 1995. Such forward-looking statements include those regarding the potential benefits of PYRUKYND® (mitapivat), tebapivat, AG-236 and AG-181; Agios’ plans, strategies and expectations for its preclinical, clinical and commercial advancement of its drug development, including PYRUKYND®, tebapivat, AG-236 and AG-181; Agios’ use of proceeds from the transaction with Royalty Pharma; potential U.S. net sales of vorasidenib and potential future royalty payments; Agios’ strategic vision and goals, including its key milestones for 2025; and the potential benefits of Agios’ strategic plans and focus. The words “anticipate,” “expect,” “goal,” “hope,” “milestone,” “plan,” “potential,” “possible,” “strategy,” “will,” “vision,” and similar expressions are intended to identify forward-looking statements, although not all forward-looking statements contain these identifying words. Such statements are subject to numerous important factors, risks and uncertainties that may cause actual events or results to differ materially from Agios’ current expectations and beliefs. For example, there can be no guarantee that any product candidate Agios is developing will successfully commence or complete necessary preclinical and clinical development phases, or that development of any of Agios’ product candidates will successfully continue. There can be no guarantee that any positive developments in Agios’ business will result in stock price appreciation. Management's expectations and, therefore, any forward-looking statements in this press release could also be affected by risks and uncertainties relating to a number of other important factors, including, without limitation: risks and uncertainties related to the impact of pandemics or other public health emergencies to Agios’ business, operations, strategy, goals and anticipated milestones, including its ongoing and planned research activities, ability to conduct ongoing and planned clinical trials, clinical supply of current or future drug candidates, commercial supply of current or future approved products, and launching, marketing and selling current or future approved products; Agios’ results of clinical trials and preclinical studies, including subsequent analysis of existing data and new data received from ongoing and future studies; the content and timing of decisions made by the U.S. FDA, the EMA or other regulatory authorities, investigational review boards at clinical trial sites and publication review bodies; Agios’ ability to obtain and maintain requisite regulatory approvals and to enroll patients in its planned clinical trials; unplanned cash requirements and expenditures; competitive factors; Agios' ability to obtain, maintain and enforce patent and other intellectual property protection for any product candidates it is developing; Agios’ ability to establish and maintain key collaborations; uncertainty regarding any royalty payments related to the sale of its oncology business or any milestone or royalty payments related to its in-licensing of AG-236, and the uncertainty of the timing of any such payments; uncertainty of the results and effectiveness of the use of Agios’ cash and cash equivalents; and general economic and market conditions. These and other risks are described in greater detail under the caption "Risk Factors" included in Agios’ public filings with the Securities and Exchange Commission . Any forward-looking statements contained in this press release speak only as of the date hereof, and Agios expressly disclaims any obligation to update any forward-looking statements, whether as a result of new information, future events or otherwise, except as required by law.
Contacts:
Investor Contact Chris Taylor , VP, Investor Relations and Corporate Communications Agios Pharmaceuticals IR@agios.com
Media Contact Eamonn Nolan , Senior Director, Corporate Communications Agios Pharmaceuticals media@agios.com
Source: Agios Pharmaceuticals, Inc.
临床3期财报临床结果申请上市高管变更
2025-04-29
WILMINGTON, Del.--(BUSINESS WIRE)--Incyte (Nasdaq:INCY) today reports 2025 first quarter financial results, and provides a status update on the Company’s clinical development portfolio.
"The double-digit revenue growth in the first quarter driven by the continued growth of Jakafi and Opzelura and the recent launch of Niktimvo, puts us on track to achieve our full year objectives," said Hervé Hoppenot, Chief Executive Officer, Incyte. "We also continued to advance our innovative pipeline, which will be critical for driving long-term growth. The positive Phase 3 results for povorcitinib in hidradenitis suppurativa in addition to the proof-of-concept in chronic spontaneous urticaria, strengthens the potential of povorcitinib as a multibillion-dollar product addressing patient needs across the five indications currently in development.”
Key Commercial Highlights
Jakafi® (ruxolitinib):
Net product revenues for the first quarter 2025 of $709 million (+24% Y/Y):
Opzelura® (ruxolitinib) cream:
Net product revenues for the first quarter 2025 of $119 million (+38% Y/Y):
Pipeline Updates
Myeloproliferative Neoplasms (MPNs) and Graft-Versus-Host Disease (GVHD) – key highlights
MPN and GVHD Programs
Indication and status
Ruxolitinib XR (QD)
(JAK1/JAK2)
Myelofibrosis, polycythemia vera and GVHD
Ruxolitinib + INCB57643
(JAK1/JAK2 + BETi)
Myelofibrosis: Phase 2
Ruxolitinib + axatilimab1
(JAK1/JAK2 + anti-CSF-1R)
Chronic GVHD: Phase 2
Steroids + axatilimab1
(Steroids + anti-CSF-1R)
Chronic GVHD: Phase 3
INCA33989
(mutCALR)
Myelofibrosis, essential thrombocythemia: Phase 1
INCB160058
(JAK2V617Fi)
Myelofibrosis: Phase 1
1 Clinical development of axatilimab in GVHD conducted in collaboration with Syndax Pharmaceuticals.
Other Hematology/Oncology – key highlights
Heme/Oncology Programs
Indication and status
Tafasitamab (Monjuvi®/Minjuvi®)
(CD19)
Relapsed or refractory diffuse large B-cell lymphoma (DLBCL): Phase 3 (B-MIND)
First-line DLBCL: Phase 3 (frontMIND)
Relapsed or refractory follicular lymphoma (FL): Phase 3 (inMIND)
Retifanlimab (Zynyz®)1
(PD-1)
Squamous cell anal cancer (SCAC): Phase 3 (POD1UM-303)
Non-small cell lung cancer (NSCLC): Phase 3 (POD1UM-304)
MSI-high endometrial cancer: Phase 2 (POD1UM-101, POD1UM-204)
INCB123667
(CDK2i)
Solid tumors with CCNE1 amplification/Cyclin E overexpression: Phase 1
INCB161734
(KRASG12D)
Advanced metastatic solid tumors with a KRASG12D mutation: Phase 1
INCA33890
(TGFßR2×PD-1)2
Advanced or metastatic solid tumors: Phase 1
1 Retifanlimab licensed from MacroGenics.
2 Development in collaboration with Merus.
Inflammation and Autoimmunity (IAI) – key highlights
Ruxolitinib Cream
Povorcitinib (INCB54707)
IAI and Dermatology Programs
Indication and status
Ruxolitinib cream (Opzelura)1
(JAK1/JAK2)
Atopic dermatitis: Phase 3 pediatric study (TRuE-AD3)
Hidradenitis suppurativa: Phase 2; Phase 3 expected to initiate in 2025
Prurigo nodularis: Phase 3 (TRuE-PN1, TRuE-PN2)
Povorcitinib
(JAK1)
Hidradenitis suppurativa: Phase 3 (STOP-HS1, STOP-HS2)
Vitiligo: Phase 3 (STOP-V1, STOP-V2)
Prurigo nodularis: Phase 3 (STOP-PN1, STOP-PN2)
Chronic spontaneous urticaria: Phase 2
Asthma: Phase 2
INCA034460
(anti-CD122)
Vitiligo: Phase 1
1 Novartis’ rights to ruxolitinib outside of the United States under our Collaboration and License Agreement with Novartis do not include topical administration.
Other - key highlights
In February 2025, Incyte and Genesis Therapeutics, Inc. (Genesis) entered into a strategic collaboration focused on the research, discovery and development of novel small molecule medicines, with an initial focus on collaboration targets selected by Incyte. Incyte receives exclusive rights to develop and commercialize collaboration products leveraged through Genesis’ GEMS artificial intelligence (AI) platform.
Other Program
Indication and Phase
Zilurgisertib
(ALK2)
Fibrodysplasia ossificans progressiva: Pivotal Phase 2
2025 First Quarter Financial Results
The financial measures presented in this press release for the three months ended March 31, 2025 and 2024 have been prepared by the Company in accordance with U.S. Generally Accepted Accounting Principles (“GAAP”), unless otherwise identified as a Non-GAAP financial measure. Management believes that Non-GAAP information is useful for investors, when considered in conjunction with Incyte’s GAAP disclosures. Management uses such information internally and externally for establishing budgets, operating goals and financial planning purposes. These metrics are also used to manage the Company’s business and monitor performance. The Company adjusts, where appropriate, for expenses in order to reflect the Company’s core operations. The Company believes these adjustments are useful to investors by providing an enhanced understanding of the financial performance of the Company’s core operations. The metrics have been adopted to align the Company with disclosures provided by industry peers.
Non-GAAP information is not prepared under a comprehensive set of accounting rules and should only be used in conjunction with and to supplement Incyte’s operating results as reported under GAAP. Non-GAAP measures may be defined and calculated differently by other companies in our industry.
As changes in exchange rates are an important factor in understanding period-to-period comparisons, Management believes the presentation of certain revenue results on a constant currency basis in addition to reported results helps improve investors’ ability to understand its operating results and evaluate its performance in comparison to prior periods. Constant currency information compares results between periods as if exchange rates had remained constant period over period. The Company calculates constant currency by calculating current year results using prior year foreign currency exchange rates and generally refers to such amounts calculated on a constant currency basis as excluding the impact of foreign exchange or being on a constant currency basis. These results should be considered in addition to, not as a substitute for, results reported in accordance with GAAP. Results on a constant currency basis, as the Company presents them, may not be comparable to similarly titled measures used by other companies and are not measures of performance presented in accordance with GAAP.
Financial Highlights
Financial Highlights
(unaudited, in thousands, except per share amounts)
Three Months Ended
March 31,
2025
2024
Total GAAP revenues
$
1,052,898
$
880,889
Total GAAP operating income
205,168
91,898
Total Non-GAAP operating income
283,641
161,183
GAAP net income
158,203
169,548
Non-GAAP net income
229,459
132,719
GAAP basic EPS
$
0.82
$
0.76
Non-GAAP basic EPS
$
1.18
$
0.59
GAAP diluted EPS
$
0.80
$
0.75
Non-GAAP diluted EPS
$
1.16
$
0.58
Revenue Details
Revenue Details
(unaudited, in thousands)
Three Months Ended
March 31,
%
Change
(as reported)
%
Change
(constant currency)1
2025
2024
Net product revenues:
Jakafi
$
709,412
$
571,839
24
%
NA
Opzelura
118,705
85,724
38
%
39
%
Iclusig
29,544
30,343
(3
%)
—
%
Pemazyre
18,440
17,676
4
%
6
%
Minjuvi/ Monjuvi
29,551
23,874
24
%
25
%
Niktimvo
13,613
—
NM
NA
Zynyz
3,009
467
544
%
NA
Total net product revenues
922,274
729,923
26
%
27
%
Royalty revenues:
Jakavi
92,145
89,595
3
%
6
%
Olumiant
30,800
30,589
1
%
6
%
Tabrecta
6,413
5,234
23
%
NA
Other
1,266
548
131
%
NM
Total royalty revenues
130,624
125,966
4
%
Total net product and royalty revenues
1,052,898
855,889
23
%
Milestone and contract revenues
—
25,000
—
%
—
%
Total GAAP revenues
$
1,052,898
$
880,889
20
%
NM = not meaningful
NA = not applicable
1 Percentage change in constant currency is calculated using 2024 foreign exchange rates to recalculate 2025 results.
Product and Royalty Revenues Total net product and royalty revenues for the quarter ended March 31, 2025 increased 23% over the prior year comparative period. Total net product revenues for the quarter ended March 31, 2025 increased 26% over the prior year comparative period primarily driven by the following:
Jakafi net product revenue increased 24% versus the prior year comparable period, driven by an increase in paid demand of 10% reflecting continued demand growth in all indications, the positive impact of the Part D redesign under the Inflation Reduction Act, partially offset by growth in 340B, and 7% favorable impact from less de-stocking compared to the first quarter of 2024. Jakafi inventory levels were within normal range at the end of the first quarter of 2025.
Opzelura net product revenue increased 38% due to continued growth in new patient starts and refills in the U.S. with U.S. paid demand up 24% versus the first quarter of 2024, partially offset by a reduction in channel inventory, and increased contribution from ex-U.S. driven by continued uptake in Germany and France, as well as growth from the recent launches in Italy and Spain. Opzelura inventory levels were within normal range at the end of the first quarter of 2025.
Minjuvi/Monjuvi net product revenue increased 24% as a result of the first quarter of 2025 reflecting three months of net product revenues in the U.S., compared to two months of net product revenue in the first quarter of 2024 due to the acquisition of U.S. rights to Monjuvi, which closed in February 2024.
Niktimvo net product revenue driven by the commercial launch of the product during the first quarter of 2025.
Operating Expenses
Operating Expense Summary
(unaudited, in thousands)
Three Months Ended
March 31,
%
Change
2025
2024
GAAP cost of product revenues
$
73,188
$
60,956
20
%
Non-GAAP cost of product revenues1
66,945
54,959
22
%
GAAP research and development
437,279
429,260
2
%
Non-GAAP research and development2
400,020
388,437
3
%
GAAP selling, general and administrative
325,691
300,256
8
%
Non-GAAP selling, general and administrative3
302,292
277,335
9
%
GAAP loss (gain) on change in fair value of acquisition-related contingent consideration
11,572
(456
)
NM
Non-GAAP loss (gain) on change in fair value of acquisition-related contingent consideration
—
—
—
%
GAAP (profit) and loss sharing under collaboration agreements
—
(1,025
)
—
%
1 Non-GAAP cost of product revenues excludes the amortization of licensed intellectual property for Iclusig relating to the acquisition of the European business of ARIAD Pharmaceuticals, Inc. and the cost of stock-based compensation.
2 Non-GAAP research and development expenses exclude the cost of stock-based compensation, MorphoSys transition costs, and Escient severance payments.
3 Non-GAAP selling, general and administrative expenses exclude the cost of stock-based compensation, MorphoSys transition costs, and Escient severance payments.
Cost of product revenues GAAP and Non-GAAP cost of product revenues for the quarter ended March 31, 2025 increased 20% and 22% respectively, compared to the same period in 2024 primarily due to increased royalty expense.
Research and development expenses GAAP and Non-GAAP research and development expense for the quarter ended March 31, 2025 increased 2% and 3%, respectively, compared to the same period in 2024, reflecting continued investment in our late stage development assets and timing of certain expenses.
Selling, general and administrative expenses GAAP and Non-GAAP selling, general and administrative expenses for the quarter ended March 31, 2025 increased 8% and 9%, respectively, compared to the same period in 2024 primarily due to timing of consumer marketing activities and of certain other expenses.
Other Financial Information
Change in fair value of acquisition-related contingent consideration The change in fair value of contingent consideration during the quarter ended March 31, 2025, compared to the same period in 2024, was primarily due to fluctuations in foreign currency exchange rates impacting future revenue projections of Iclusig.
Operating income GAAP and Non-GAAP operating income for the three months ended March 31, 2025 increased 123% and 76%, respectively, compared to the same period in 2024, driven primarily by growth in net product revenue.
Cash, cash equivalents and marketable securities position As of March 31, 2025 and December 31, 2024, cash, cash equivalents and marketable securities totaled $2.4 billion and $2.2 billion, respectively.
2025 Financial Guidance
Incyte's guidance for the fiscal year 2025 is summarized below. Incyte is raising its full year 2025 Jakafi revenue guidance. Guidance for Opzelura includes net product revenue for pediatric atopic dermatitis which has an anticipated approval in the second half of 2025. Guidance for other oncology net product revenues include net product revenue for Monjuvi in follicular lymphoma and Zynyz in squamous cell anal carcinoma. Approvals for these indications are anticipated in the second half of 2025. Guidance for research and development excludes the $15 million of expense for the full year 2025 related to our recently announced collaboration with Genesis.
Current
Previous
Jakafi net product revenues
$2,950 - $3,000 million
$2,925 - $2,975 million
Opzelura net product revenues
Unchanged
$630 - $670 million
Other oncology net product revenues(1)
Unchanged
$415 - $455 million
GAAP Cost of product revenues
Unchanged
8.5% - 9.0% of net product revenues
Non-GAAP Cost of product revenues(2)
Unchanged
7.5% - 8.0% of net product revenues
GAAP Research and development expenses
Unchanged
$1,930 - $1,960 million
Non-GAAP Research and development expenses(3)
Unchanged
$1,780 - $1,805 million
GAAP Selling, general and administrative expenses
Unchanged
$1,280 - $1,310 million
Non-GAAP Selling, general and administrative expenses(3)
Unchanged
$1,160 - $1,185 million
1Pemazyre in the U.S., EU and Japan; Niktimvo, Monjuvi and Zynyz in the U.S.; and Iclusig and Minjuvi in the EU.
2Adjusted to exclude the amortization of licensed intellectual property for Iclusig relating to the acquisition of the European business of ARIAD Pharmaceuticals, Inc. and the estimated cost of stock-based compensation.
3Adjusted to exclude the estimated cost of stock-based compensation.
Conference Call and Webcast Information
Incyte will hold a conference call and webcast this morning at 8:00 a.m. ET. To access the conference call, please dial 877-407-3042 for domestic callers or 201-389-0864 for international callers. When prompted, provide the conference identification number, 13753168.
If you are unable to participate, a replay of the conference call will be available for 90 days. The replay dial-in number for the United States is 877-660-6853 and the dial-in number for international callers is 201-612-7415. To access the replay you will need the conference identification number, 13753168.
The conference call will also be webcast live and can be accessed at investor.incyte.com.
About Incyte
A global biopharmaceutical company on a mission to Solve On., Incyte follows the science to find solutions for patients with unmet medical needs. Through the discovery, development and commercialization of proprietary therapeutics, Incyte has established a portfolio of first-in-class medicines for patients and a strong pipeline of products in Oncology and Inflammation & Autoimmunity. Headquartered in Wilmington, Delaware, Incyte has operations in North America, Europe and Asia.
For additional information on Incyte, please visit Incyte.com or follow us on social media: LinkedIn, X, Instagram, Facebook, YouTube.
About Jakafi® (ruxolitinib)
Jakafi® (ruxolitinib) is a JAK1/JAK2 inhibitor approved by the U.S. FDA for treatment of polycythemia vera (PV) in adults who have had an inadequate response to or are intolerant of hydroxyurea; intermediate or high-risk myelofibrosis (MF), including primary MF, post-polycythemia vera MF and post-essential thrombocythemia MF in adults; steroid-refractory acute GVHD in adult and pediatric patients 12 years and older; and chronic GVHD after failure of one or two lines of systemic therapy in adult and pediatric patients 12 years and older.
Jakafi is a registered trademark of Incyte.
About Opzelura® (ruxolitinib) Cream
Opzelura® (ruxolitinib) cream, a novel cream formulation of Incyte’s selective JAK1/JAK2 inhibitor ruxolitinib, approved by the U.S. Food & Drug Administration for the topical treatment of nonsegmental vitiligo in patients 12 years of age and older, is the first and only treatment for repigmentation approved for use in the United States. Opzelura is also approved in the U.S. for the topical short-term and non-continuous chronic treatment of mild to moderate atopic dermatitis (AD) in non-immunocompromised patients 12 years of age and older whose disease is not adequately controlled with topical prescription therapies, or when those therapies are not advisable. Use of Opzelura in combination with therapeutic biologics, other JAK inhibitors, or potent immunosuppressants, such as azathioprine or cyclosporine, is not recommended.
In Europe, Opzelura (ruxolitinib) cream 15mg/g is approved for the treatment of non-segmental vitiligo with facial involvement in adults and adolescents from 12 years of age.
Incyte has worldwide rights for the development and commercialization of ruxolitinib cream, marketed in the United States as Opzelura.
Opzelura and the Opzelura logo are registered trademarks of Incyte.
About Monjuvi® (tafasitamab-cxix)
Monjuvi® (tafasitamab-cxix) is a humanized Fc-modified cytolytic CD19 targeting monoclonal antibody. In 2010, MorphoSys licensed exclusive worldwide rights to develop and commercialize tafasitamab from Xencor, Inc. Tafasitamab incorporates an XmAb® engineered Fc domain, which mediates B-cell lysis through apoptosis and immune effector mechanism including Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) and Antibody-Dependent Cellular Phagocytosis (ADCP). MorphoSys and Incyte entered into: (a) in January 2020, a collaboration and licensing agreement to develop and commercialize tafasitamab globally; and (b) in February 2024, an agreement whereby Incyte obtained exclusive rights to develop and commercialize tafasitamab globally.
Following accelerated approval by the U.S. Food and Drug Administration in July 2020, Monjuvi® (tafasitamab-cxix) is being commercialized in the United States by Incyte. In Europe, Minjuvi® (tafasitamab) received conditional Marketing Authorization from the European Medicines Agency in August 2021.
XmAb® is a registered trademark of Xencor, Inc.
Monjuvi, Minjuvi, the Minjuvi and Monjuvi logos and the “triangle” design are (registered) trademarks of Incyte.
About Pemazyre® (pemigatinib)
Pemazyre® (pemigatinib) is a kinase inhibitor indicated in the United States for the treatment of adults with previously treated, unresectable locally advanced or metastatic cholangiocarcinoma with a fibroblast growth factor receptor 2 (FGFR2) fusion or other rearrangement as detected by an FDA-approved test. This indication is approved under accelerated approval based on overall response rate and duration of response. Continued approval for this indication may be contingent upon verification and description of clinical benefit in a confirmatory trial(s).
Pemazyre is also the first targeted treatment approved for use in the United States for treatment of adults with relapsed or refractory myeloid/lymphoid neoplasms (MLNs) with FGFR1 rearrangement.
In Japan, Pemazyre is approved for the treatment of patients with unresectable biliary tract cancer (BTC) with a fibroblast growth factor receptor 2 (FGFR2) fusion gene, worsening after cancer chemotherapy.
In Europe, Pemazyre is approved for the treatment of adults with locally advanced or metastatic cholangiocarcinoma with a fibroblast growth factor receptor 2 (FGFR2) fusion or rearrangement that have progressed after at least one prior line of systemic therapy.
Pemazyre is a potent, selective, oral inhibitor of FGFR isoforms 1, 2 and 3 which, in preclinical studies, has demonstrated selective pharmacologic activity against cancer cells with FGFR alterations.
Pemazyre is marketed by Incyte in the United States, Europe and Japan.
Pemazyre is a trademark of Incyte.
About Iclusig® (ponatinib) tablets
Iclusig® (ponatinib) targets not only native BCR-ABL but also its isoforms that carry mutations that confer resistance to treatment, including the T315I mutation, which has been associated with resistance to other approved TKIs.
In the EU, Iclusig is approved for the treatment of adult patients with chronic phase, accelerated phase or blast phase chronic myeloid leukemia (CML) who are resistant to dasatinib or nilotinib; who are intolerant to dasatinib or nilotinib and for whom subsequent treatment with imatinib is not clinically appropriate; or who have the T315I mutation, or the treatment of adult patients with Philadelphia-chromosome positive acute lymphoblastic leukemia (Ph+ ALL) who are resistant to dasatinib; who are intolerant to dasatinib and for whom subsequent treatment with imatinib is not clinically appropriate; or who have the T315I mutation.
Click here to view the Iclusig EU Summary of Medicinal Product Characteristics.
Incyte has an exclusive license from Takeda Pharmaceuticals International AG to commercialize ponatinib in the European Union and 29 other countries, including Switzerland, UK, Norway, Turkey, Israel and Russia. Iclusig is marketed in the U.S. by Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited.
About Zynyz® (retifanlimab-dlwr)
Zynyz® (retifanlimab) is an intravenous PD-1 inhibitor indicated in the U.S. for the treatment of adult patients with metastatic or recurrent locally advanced Merkel cell carcinoma (MCC). This indication is approved under accelerated approval based on tumor response rate and duration of response. Continued approval for this indication may be contingent upon verification and description of clinical benefit in confirmatory trials.
Zynyz is marketed by Incyte in the U.S. In 2017, Incyte entered into an exclusive collaboration and license agreement with MacroGenics, Inc. for global rights to retifanlimab.
Zynyz is a trademark of Incyte.
About Niktimvo™ (axatilimab-csfr)
Niktimvo (axatilimab-csfr) is a first-in-class colony stimulating factor-1 receptor (CSF-1R)-blocking antibody approved for use in the U.S. for the treatment of chronic graft-versus-host disease (GVHD) after failure of at least two prior lines of systemic therapy in adult and pediatric patients weighing at least 40 kg (88.2 lbs).
In 2016, Syndax licensed exclusive worldwide rights to develop and commercialize axatilimab from UCB. In September 2021, Syndax and Incyte entered into an exclusive worldwide co-development and co-commercialization license agreement for axatilimab in chronic GVHD and any future indications.
Axatilimab is being studied in frontline combination trials in chronic GVHD – a Phase 2 combination trial with ruxolitinib (NCT06388564) and a Phase 3 combination trial with steroids (NCT06585774) are underway. Axatilimab is also being studied in an ongoing Phase 2 trial in patients with idiopathic pulmonary fibrosis (NCT06132256).
Niktimvo is a trademark of Incyte.
All other trademarks are the property of their respective owners.
Forward-Looking Statements
Except for the historical information set forth herein, the matters set forth in this release contain predictions, estimates and other forward-looking statements, including any discussion of the following: Incyte’s potential for continued performance and growth; Incyte’s ability to achieve both its full-year and long-term objectives; Incyte’s financial guidance for 2025, including its expectations regarding sales of and demand for Jakafi and Opzelura; expected revenue contribution from Niktimvo and additional near-term launches; the potential of povorcitinib to be a multibillion-dollar product; the possibility for 2025 to be a transformational year for Incyte in terms of launches, phase 3 study initiations, pivotal readouts and proof of concept readouts; Incyte’s potential to have more than 10 high impact launches by 2030; the potential and progress of programs in our pipeline; ongoing clinical trials and clinical trials that may be initiated; expectations regarding discussions with regulators, regulatory submissions and regulatory approvals; plans to present data at upcoming medical conferences; Incyte’s exposure to potential tariffs; and 2025 newsflow items.
These forward-looking statements are based on Incyte’s current expectations and subject to risks and uncertainties that may cause actual results to differ materially, including unanticipated developments in and risks related to: further research and development and the results of clinical trials possibly being unsuccessful or insufficient to meet applicable regulatory standards or warrant continued development; the ability to enroll sufficient numbers of subjects in clinical trials and the ability to enroll subjects in accordance with planned schedules; determinations made by the FDA, EMA and other regulatory agencies; Incyte’s dependence on its relationships with and changes in the plans of its collaboration partners; the efficacy or safety of Incyte’s products and the products of Incyte’s collaboration partners; the acceptance of Incyte’s products and the products of Incyte’s collaboration partners in the marketplace; market competition; unexpected variations in the demand for Incyte’s products and the products of Incyte’s collaboration partners; the effects of announced or unexpected price regulation or limitations on reimbursement or coverage for Incyte’s products and the products of Incyte’s collaboration partners; sales, marketing, manufacturing and distribution requirements, including Incyte’s and its collaboration partners’ ability to successfully commercialize and build commercial infrastructure for newly approved products and any additional products that become approved; greater than expected expenses, including expenses relating to litigation or strategic activities; variations in foreign currency exchange rates; and other risks detailed in Incyte’s reports filed with the Securities and Exchange Commission, including its annual report on form 10-K for the year ended December 31, 2024. Incyte disclaims any intent or obligation to update these forward-looking statements.
INCYTE CORPORATION
CONDENSED CONSOLIDATED STATEMENTS OF OPERATIONS
(unaudited, in thousands, except per share amounts)
Three Months Ended
March 31,
2025
2024
GAAP
Revenues:
Product revenues, net
$
922,274
$
729,923
Product royalty revenues
130,624
125,966
Milestone and contract revenues
—
25,000
Total revenues
1,052,898
880,889
Costs, expenses and other:
Cost of product revenues (including definite-lived intangible amortization)
73,188
60,956
Research and development
437,279
429,260
Selling, general and administrative
325,691
300,256
Loss (gain) on change in fair value of acquisition-related contingent consideration
11,572
(456
)
(Profit) and loss sharing under collaboration agreements
—
(1,025
)
Total costs, expenses and other
847,730
788,991
Income from operations
205,168
91,898
Interest income
22,929
46,770
Interest expense
(660
)
(430
)
(Loss) gain on equity investments
(1,343
)
99,947
Other, net
8,096
(2,026
)
Income before provision for income taxes
234,190
236,159
Provision for income taxes
75,987
66,611
Net income
$
158,203
$
169,548
Net income per share:
Basic
$
0.82
$
0.76
Diluted
$
0.80
$
0.75
Shares used in computing net income per share:
Basic
193,712
224,484
Diluted
198,197
227,219
INCYTE CORPORATION
CONDENSED CONSOLIDATED BALANCE SHEETS
(unaudited, in thousands)
March 31,
2025
December 31,
2024
ASSETS
Cash, cash equivalents and marketable securities
$
2,408,658
$
2,158,092
Accounts receivable
823,134
853,154
Property and equipment, net
765,359
763,411
Finance lease right-of-use assets, net
29,892
30,803
Inventory
429,286
407,199
Prepaid expenses and other assets
243,470
181,382
Equity investments
17,463
18,814
Other intangible assets, net
107,611
113,803
Goodwill
155,593
155,593
Deferred income tax asset
768,899
762,071
Total assets
$
5,749,365
$
5,444,322
LIABILITIES AND STOCKHOLDERS’ EQUITY
Accounts payable, accrued expenses and other liabilities
$
1,849,681
$
1,765,733
Finance lease liabilities
37,121
37,961
Acquisition-related contingent consideration
195,000
193,000
Stockholders’ equity
3,667,563
3,447,628
Total liabilities and stockholders’ equity
$
5,749,365
$
5,444,322
INCYTE CORPORATION
RECONCILIATION OF GAAP NET (LOSS) INCOME TO SELECTED NON-GAAP ADJUSTED INFORMATION
(unaudited, in thousands, except per share amounts)
Three Months Ended
March 31,
2025
2024
GAAP Net Income
$
158,203
$
169,548
Adjustments1:
Non-cash stock compensation from equity awards (R&D)2
36,724
36,792
Non-cash stock compensation from equity awards (SG&A)2
23,399
22,373
Non-cash stock compensation from equity awards (COGS)2
859
613
Non-cash interest3
82
108
Loss (gain) on equity investments4
1,343
(99,947
)
Amortization of acquired product rights5
5,384
5,384
Loss (gain) on change in fair value of contingent consideration6
11,572
(456
)
MorphoSys transition costs7
—
4,579
Escient acquisition related compensation expense8
535
—
Tax effect of Non-GAAP pre-tax adjustments9
(8,642
)
(6,275
)
Non-GAAP Net Income
$
229,459
$
132,719
Non-GAAP net income per share:
Basic
$
1.18
$
0.59
Diluted
$
1.16
$
0.58
Shares used in computing Non-GAAP net income per share:
Basic
193,712
224,484
Diluted
198,197
227,219
1 Included within the Milestone and contract revenues line item in the Condensed Consolidated Statements of Operations (in thousands) for the three months ended March 31, 2025 are milestones of $0 earned from our collaborative partners, as compared to $25,000 of milestones earned for the three months ended March 31, 2024. Included within the Research and development expenses line item in the Condensed Consolidated Statements of Operations (in thousands) for the three months ended March 31, 2025 and 2024 are upfront consideration and milestones of $15,500 and $1,000, respectively, related to our collaborative partners.
2 As included within the Cost of product revenues (including definite-lived intangible amortization) line item; the Research and development expenses line item; and the Selling, general and administrative expenses line item in the Condensed Consolidated Statements of Operations.
3 As included within the Interest expense line item in the Condensed Consolidated Statements of Operations.
4 As included within the (Loss) gain on equity investments line item in the Condensed Consolidated Statements of Operations.
5 As included within the Cost of product revenues (including definite-lived intangible amortization) line item in the Condensed Consolidated Statements of Operations. Acquired product rights of licensed intellectual property for Iclusig is amortized utilizing a straight-line method over the estimated useful life of 12.5 years.
6 As included within the Loss (gain) on change in fair value of acquisition-related contingent consideration line item in the Condensed Consolidated Statements of Operations.
7 Included within the Research and development line item in the Condensed Consolidated Statements of Operations (in thousands) is $4,031 for the three months ended March 31, 2024, and $548 is included within the Selling, general and administrative expenses line item in the Condensed Consolidated Statements of Operations (in thousands) for the three months ended March 31, 2024. MorphoSys transition costs primarily represent employee related costs to transition research and development and selling, general and administrative activities to us under the former collaboration agreement with MorphoSys.
8 Included within the Research and development line item in the Condensed Consolidated Statements of Operations (in thousands) is $535 for the three months ended March 31, 2025. Escient acquisition related compensation expense represents non-recurring charges associated with severance payments to former Escient employees.
9 Income tax effects of Non-GAAP pre-tax adjustments are calculated using an estimated annual effective tax rate, taking into consideration any permanent items and valuation allowances against related deferred tax assets. The Non-GAAP net income for the three months ended March 31, 2024 should have been $132,719 compared to the $145,269 previously disclosed to correct a transposition error in the tax effect of Non-GAAP pre-tax adjustments. For the three months ended March 31, 2024, the tax effect of Non-GAAP pre-tax adjustments should have been ($6,275) instead of $6,275. This correction is reflected in the table above.
临床1期临床3期上市批准引进/卖出财报
2025-04-29
·药智网
美国临床肿瘤学会(ASCO)年会是全球规模最大、学术水平最高、最具权威性的临床肿瘤学会议,会议汇集了众多世界一流的肿瘤学专家、患者倡导者和工业界代表,共同分享探讨当前国际最前沿的临床肿瘤学科研成果和肿瘤治疗技术,推动肿瘤学领域的科学进步。今年的ASCO年会将于5月30日到6月3日在芝加哥举办。据官方统计,本次ASCO年会预计开展场次超200场,涵盖24场口头摘要专场(Oral Abstract Session)、2700篇壁报(Poster)展示。国内药企对ASCO并不陌生,很多公司都是常客。今年也不例外,众多国内药企积极参与,展示了其在新药研发方面的活跃度和竞争力,包括恒瑞医药、正大天晴、信达生物、泽璟制药、映恩生物、维立志博、亚盛医药、荣昌生物、迪哲医药、科伦博泰等公司均有新药研究入选。泽璟制药:将公布28项最新临床研究数据,这些数据将进一步展现早期抗体管线在更大人群中的有效性及安全性。主要包括CD3/DLL3/DLL3三特异性抗体单药治疗晚期小细胞肺癌的Ⅱ期剂量扩展研究,PD-1/TIGIT双特异性抗体ZG005治疗一线以及二线以上宫颈癌的拓展临床数据,LAG-3/TIGIT双特异性抗体ZGGS15单药治疗晚期实体瘤患者的人体研究等。映恩生物:将以口头报告形式公布两项研究数据:HER3 ADC药物DB-1310用于晚期实体瘤患者的1/2a期研究初步结果和B7H3 ADC药物DB-1311/BNT324用于经多线治疗的去势抵抗性前列腺癌(CRPC)患者的数据。维立志博:将以口头报告形式公布PD-L1/4-1BB双特异性抗体LBL-024一线治疗晚期肺外神经内分泌癌(EP-NEC)的突破性临床数据。以及以壁报形式展示LAG-3抗体LBL-007联合PD-1单抗和化疗一线治疗复发性或转移性鼻咽癌(R/M NPC)的2期研究最新数据。信达生物:7项研究入选ASCO口头报告,包括PD-1/IL-2α-bias双特异性抗体融合蛋白IBI363三个不同适应症(NSCLC、CRC、MLN)的临床数据,CLDN18.2 ADC药物IBI343在胰腺导管腺癌(PDAC)中的I期剂量扩展队列研究结果,以及PD-1单抗的三项临床研究成果。除此之外,IBI354(HER2 ADC)、IBI130(TROP2 ADC)等分子也将在此次会议中公布临床数据。正大天晴:有12项口头报告,加上壁报和摘要收录等形式,共有40多项创新成果将在大会公布最新临床数据。例如:贝莫苏拜单抗联合安罗替尼对比帕博利珠单抗一线治疗晚期非小细胞肺癌的Ⅲ期研究、STUPP方案联合或不联合安罗替尼治疗新诊断胶质母细胞瘤的Ⅱ期临床试验结果、安罗替尼对比贝伐珠单抗联合化疗一线治疗RAS/BRAF野生型不可切除转移性结直肠癌的Ⅲ期临床研究(ANCHOR研究)结果。荣昌生物:有20余项肿瘤管线临床进展或阶段性数据入选口头报告、壁报或线上展示,其中包括维迪西妥单抗联合疗法一线治疗HER2表达胃癌、维迪西妥单抗联合RC148(PD-1/VEGF双抗)一线治疗三阴性乳腺癌、RC108(c-Met ADC)联合TKI二线治疗EFGR突变伴MET过表达非小细胞肺癌等临床研究。恒瑞医药:有多款ADC、双抗药物入选,包括SHR-1826(c-MET ADC)、SHR-A2102(Nectin-4 ADC)、SHR-A1811(HER2 ADC)、SHR-A1912(CD79b ADC)、SHR-1701(PD-L1/TGF-β双抗)等,覆盖了多种肿瘤类型,展现了其在肿瘤治疗领域的强大研发管线和创新能力。科伦博泰:6项研究成果将于ASCO发布,涉及TROP2 ADC芦康沙妥珠单抗(sac-TMT)、PD-L1单抗(A167)以及RET抑制剂KL590586(A400/EP0031)三款药物,其中SKB263的OptiTROP-Lung03临床(EGFRm NSCLC 3L)和OptiTROP-Breast05临床(mTNBC 1L),以及A167的NPC 1L的ph3临床数据获得了口头报告。并且,SKB264还将更新联合A167治疗nsqNSCLC 1L患者的ph2临床数据(OptiTROP-Lung01)。亚盛医药:核心品种Bcl-2选择性抑制剂Lisaftoclax(APG-2575)斩获口头报告,将公布APG-2575联合疗法在初治(TN)或既往接受过维奈克拉治疗的髓系恶性肿瘤患者中的最新进展。另外亚盛医药将展示MDM2-p53抑制剂Alrizomadlin(APG-115)治疗晚期腺样囊性癌(ACC)或其他实体瘤患者的最新临床数据。迪哲医药:将在本次ASCO会议发布3项研究成果,其中DZD8586(BTK/LYN双靶点抑制剂)后线治疗CLL/SLL的ph1/2临床获得口头报告,DZD6008(四代EGFR TKI)也将首次发表临床数据。结语:国产新药在2025年ASCO大会上的展示反映了国内医药研发的蓬勃发展态势,在肿瘤治疗领域不断取得新的进展和突破,为全球患者带来了更多希望和选择。未来,随着这些新药研发的不断推进,有望进一步提升国产药物在全球医药市场的竞争力和影响力。参考来源:各公司公开资料声明:本内容仅用作医药行业信息传播,不代表药智网立场。对本文有异议或投诉,请联系maxuelian@yaozh.com。责任编辑 | 小月石合作、投稿 | 马老师 18323856316(同微信) 阅读原文,是受欢迎的文章哦
ASCO会议抗体药物偶联物临床结果临床1期临床2期
分析
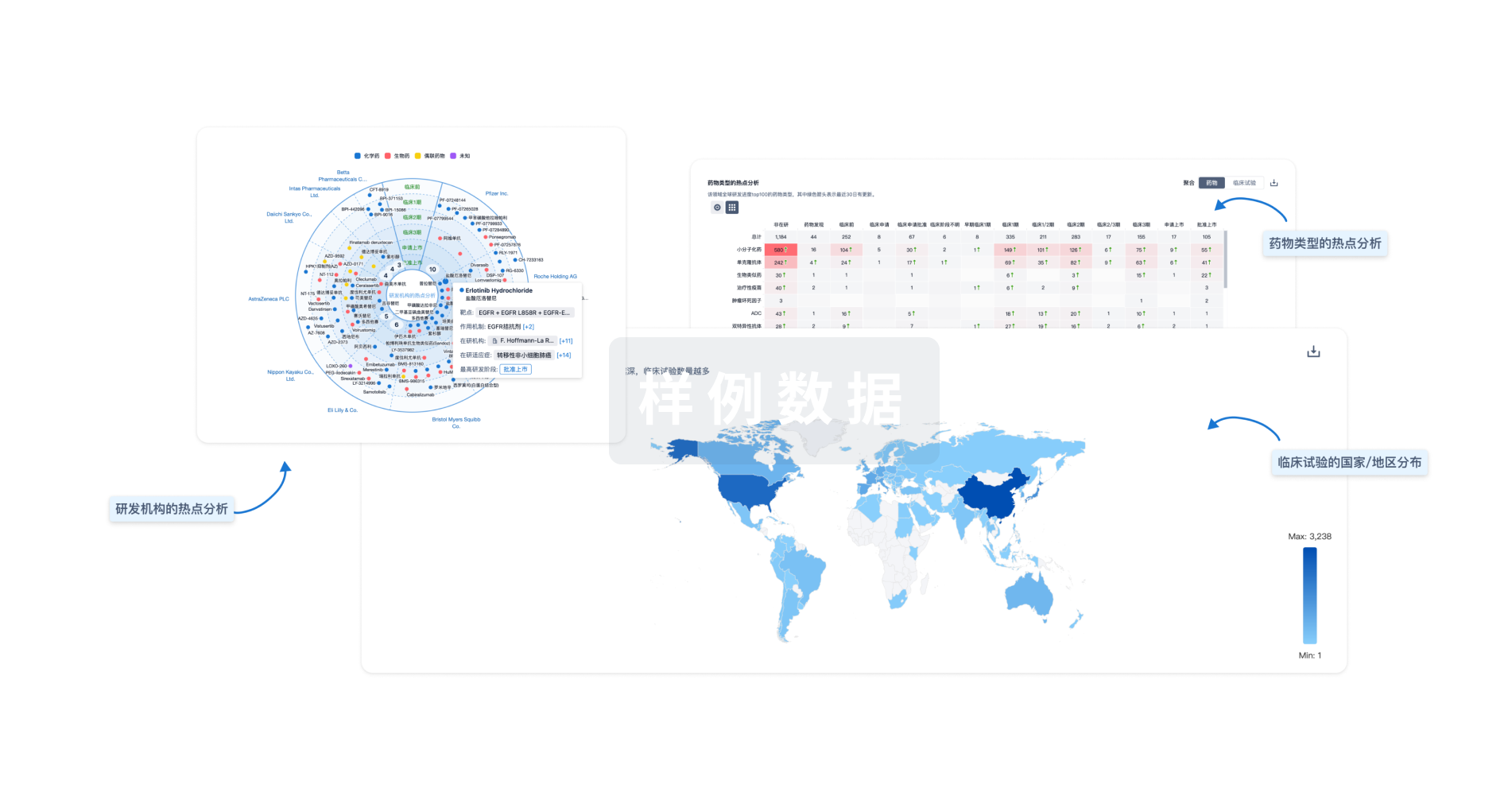
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用