预约演示
更新于:2025-05-07
HLA class II抗原
更新于:2025-05-07
基本信息
别名 Major histocompatibility complex II、主要组织相容性复合体II |
简介- |
关联
39
项与 HLA class II抗原 相关的药物作用机制 HLA class I抗原调节剂 [+2] |
在研机构 |
原研机构 |
在研适应症 |
非在研适应症- |
最高研发阶段申请上市 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
作用机制 HLA class II抗原调节剂 [+3] |
原研机构 |
最高研发阶段临床3期 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
作用机制 HLA class II抗原调节剂 [+3] |
在研机构 |
最高研发阶段临床2期 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
51
项与 HLA class II抗原 相关的临床试验CTRI/2025/03/082306
TACTI-004, a double-blinded, randomized phase 3 trial in patients with advanced/metastatic non-small cell lung cancer (NSCLC) receiving eftilagimod alfa (MHC class II agonist) in combination with pembrolizumab (PD-1 antagonist) and chemotherapy. - TACTI-004 (IMP321-P026) ; Keynote-F91
开始日期2025-03-31 |
申办/合作机构 |
NCT06726265
TACTI-004, a Double-Blinded, Randomized Phase 3 Trial in Patients With Advanced/Metastatic Non-Small Cell Lung Cancer (NSCLC) Receiving Eftilagimod Alfa (MHC Class II Agonist) in Combination With Pembrolizumab (PD-1 Antagonist) and Chemotherapy.
The purpose of this study is to compare eftilagimod alfa (efti) in combination with pembrolizumab and chemotherapy versus placebo in combination with pembrolizumab and chemotherapy with respect to overall survival (OS) and progression-free survival (PFS) per Response Evaluation Criteria in Solid Tumors Version 1.1 (RECIST 1.1) among adults with metastatic non-small cell lung cancer (NSCLC).
Participants will receive either efti plus standard treatment (pembrolizumab and platinum doublet chemotherapy) or placebo plus standard treatment and will be treated for up to 2 years.
Participants will receive either efti plus standard treatment (pembrolizumab and platinum doublet chemotherapy) or placebo plus standard treatment and will be treated for up to 2 years.
开始日期2025-03-21 |
申办/合作机构 Immutep SAS [+1] |
ACTRN12624000316505
A phase IC, randomized, double-blind, placebo-controlled, multicenter study to evaluate the safety, pharmacokinetics, immunogenicity, and biological effects of DONQ52 in celiac disease patients with gluten challenge
开始日期2024-07-16 |
100 项与 HLA class II抗原 相关的临床结果
登录后查看更多信息
100 项与 HLA class II抗原 相关的转化医学
登录后查看更多信息
0 项与 HLA class II抗原 相关的专利(医药)
登录后查看更多信息
33,070
项与 HLA class II抗原 相关的文献(医药)2025-12-31·Emerging Microbes & Infections
Vδ2 T-cells response in people with Mpox infection: a three-month longitudinal assessment
Article
作者: Maggi, Fabrizio ; Oliva, Alessandra ; Coppola, Andrea ; Mondi, Annalisa ; Fontana, Carla ; Notari, Stefania ; Messina, Francesco ; Antinori, Andrea ; Tempestilli, Massimo ; Cimini, Eleonora ; Prota, Gianluca ; Girardi, Enrico ; Matusali, Giulia ; Mazzotta, Valentina ; Tartaglia, Eleonora
2025-12-31·Epigenetics
Differential methylation in blood pressure control genes is associated to essential hypertension in African Brazilian populations
Article
作者: Magalhães Borges, Vinícius ; Maschietto, Mariana ; Victorino Krepischi, Ana Cristina ; Alvizi, Lucas ; Avila Martins, Camila Cristina ; Nunes, Kelly ; Mingroni-Netto, Regina Célia ; Kimura, Lilian
2025-12-31·OncoImmunology
Molecular and immunological features associated with long-term benefits in metastatic NSCLC patients undergoing immune checkpoint blockade
Article
作者: Navarro-Gorro, Nil ; Del Rey-Vergara, Raúl ; Berenguer-Molins, Pau ; White, James ; Perera-Bel, Júlia ; Arriola, Edurne ; Jackson, Jennifer B ; Masfarré, Laura ; Hardy-Werbin, Max ; Rocha, Pedro ; Lu, Wei ; Sausen, Mark ; Taus, Álvaro ; Clavé, Sergi ; Galindo, Miguel ; Rovira, Ana ; Chalela, Roberto ; Hernandez, Sharia ; Villanueva, Xavier ; Soto, Luisa M Solis ; Bach, Rafael ; Iñañez, Albert ; Sanchéz, Ignacio ; Acedo-Terrades, Ariadna ; Curull, Victor ; Bellosillo, Beatriz ; Sanchéz-Espiridion, Beatriz ; Rossell, Adrià ; Giner, Mario ; Sánchez-Font, Albert ; Georgiadis, Andrew ; Moliner, Laura ; Wistuba, Ignacio
106
项与 HLA class II抗原 相关的新闻(医药)2025-05-02
·抗体圈
这篇文章于2025年发表在Stem Cell Reviews and Reports,全面评估了临床级MSC可扩展生产的自动化制造工艺和平台。摘要: 间充质干细胞/基质细胞(MSC)具有卓越的再生和免疫调节特性,因此在再生医学方面具有巨大的潜力。然而,它们的治疗应用需要在严格的监管标准和良好生产规范(GMP)指南下进行大规模生产,这带来了重大挑战。本综述全面评估了临床级MSC可扩展生产的自动化制造工艺和平台。分析了各种大型培养容器,包括多层培养瓶和生物反应器,它们在MSC扩增中的功效。此外,还回顾和比较了自动化MSCs生产平台,如Quantum®细胞扩增系统、CliniMACSProdigy®、NANT001/XL、CellQualia™、Cocoon®平台和Xuri™细胞扩增系统W25。我们还强调了优化培养基的重要性,特别强调从胎牛血清转向人源化或无血清替代品,以满足GMP标准。此外,还讨论了替代冷冻保存方法和控速冷冻系统的进展,这些方法为MSC保存提供了有希望的改进。总之,推进自动化制造工艺和平台对于实现基于MSC的再生医学的全部潜力和满足对基于细胞的疗法日益增长的需求至关重要。强调涉及行业、学术界和监管机构的合作举措,以加速基于MSC的疗法向临床实践的转化。1.背景介绍 间充质干细胞/基质细胞(MSC)是一种存在于各种组织中的多能成体干细胞,包括骨髓、脂肪组织和脐带。它们具有显着的自我更新和分化能力,使它们能够产生各种细胞类型,如成骨细胞、软骨细胞或脂肪细胞。此外,MSC分泌多种生物活性分子,包括生长因子、细胞因子和细胞外囊泡,有助于组织修复和再生。这些旁分泌因子发挥抗炎、抗凋亡和促血管生成作用,促进各种损伤和疾病的组织愈合。此外,使MSC对治疗应用具有吸引力的关键特征之一是其免疫调节特性。MSC已被证明可以通过抑制各种免疫细胞(包括T细胞、B细胞、巨噬细胞和树突状细胞)的增殖和功能来抑制免疫反应。这种免疫调节作用使MSC成为治疗各种自身免疫性疾病、移植物抗宿主病(GVHD)和炎症综合征的有希望的候选者。因此,MSC为再生医学、组织工程和自身免疫性疾病的治疗提供了一条有前途的途径,因此在解决未满足的医疗需求方面具有巨大潜力。 由于MSC的生产被视为ATMP(先进疗法药品),因此其应用由特定的监管框架控制。MSC生产需要符合当前良好生产规范(GMP)标准的大规模细胞扩增系统。GMP指南是由政府监管机构制定的严格法规,例如美国的食品药品监督管理局(FDA)或欧洲药品管理局(EMA),旨在确保药品(包括基于MSC的产品)的质量、安全性和有效性。考虑到基于MSC的疗法涉及事后无法过滤或灭菌的活细胞的给药,MSC生产的首要GMP先决条件是具有受控无菌和多级无菌保护解决方案的环境。洁净室设施、无菌防护服、层流罩和封闭培养系统共同参与其中,以限制空气中的颗粒数量并避免最终产品受到污染。最后但并非最不重要的一点是,训练有素、组织有序、了解制造过程的员工对于不伤害患者也很重要。 基于MSC的疗法的生产过程包括供体和MSC来源选择、细胞分离和扩增、质量控制以及扩增细胞的进一步移植等步骤。将MSC用于治疗目的的关键挑战之一是能够在保持其特性和功能特性的同时大量扩增它们。一般来说,天然组织和收集的生物材料中MSCs的初始频率非常低——据报道,骨髓中每104-105个单核细胞或脂肪抽吸物中每102-103个细胞中MSC少于或大约1个。传统的MSC体外扩增至临床相关产量(数百万至数亿个细胞)通常非常耗时、劳动密集型,并且需要大量的培养箱空间。因此,它不仅效率低下,而且还可能损害扩增的MSC的质量。重要的是,以前的研究报道了MSC在晚期传代中的增殖和分化能力下降。 MSC制造过程的另一个组成部分是最终产品的质量控制。这包括细胞活力、表型标志物、分化潜能和无微生物污染的检测,以评估制造细胞产品的纯度、特性、效力和安全性。根据国际细胞治疗学会(ISCT)的规定,MSC必须满足以下方面:(I)塑性粘附性,(II)CD105、CD73和CD90等分化簇(CD)分子表达阳性,缺乏CD45、CD34、CD14、CD11b、CD79α和人类白细胞抗原(HLA)-DR的表达,(III)在体外分化为脂肪细胞、软骨细胞和成骨细胞[9].然而,为了确保MSC的安全性和功能性,应进行免疫调节活性和基因组稳定性的额外测试。2.MSC生产平台 大规模生产临床级MSC的生物工艺策略是一个复杂的过程,对封闭和无菌环境有很高的要求。因此,MSC加工必须在符合GMP的专用设施中进行,以最大限度地降低物理屏障和空气过滤污染的风险。为了响应该领域的客户需求,一些制造商正在开发和引入用于自动和密闭细胞培养的新技术,例如量子®细胞扩增系统(TerumoBCT)、CliniMACSProdigy®(MiltenyiBiotec)、NANT001/NANTXL系统(VivaBioCell)、CellQualia™(Sinfonia技术)、Cocoon®平台(Lonza)或Xuri™细胞扩增系统W25(Cytiva)(图1).生物反应器的利用在扩大基于细胞的疗法的可及性方面具有巨大潜力,特别是对于缺乏现场GMP合规实验室的中心。这些生物反应器为细胞培养和扩增提供了受控的环境,确保了细胞生产过程的一致性、可重复性和质量。通过使用生物反应器,没有GMP设施的中心仍然可以生产临床级细胞,从而将基于细胞的治疗范围扩大到更多患者。图1:大规模生产临床级间充质干细胞/基质细胞(MSC)的生物加工策略的复杂概述。MSC扩增平台(Quantum®细胞扩增系统、CliniMACSProdigy®、NANT001/XL、CellQualia™、Cocoon®平台和Xuri™细胞扩增系统W25)需要以下输入:样品或分离的细胞、培养基、洗涤试剂(磷酸盐缓冲盐水)、解离试剂(胰蛋白酶、TrypLE)和可选添加剂。细胞收获、质量控制(最终产品的纯度、特性、效力和安全性)、冷冻保存和产品配方等下游工艺也是临床级MSC生产的重要组成部分。 Terumo-BCT开发的量子®细胞扩增系统代表了一种利用中空纤维生物反应器的自动和闭式细胞培养的替代策略。此外,该设备以适合细胞培养过程的速率提供连续的培养基更换。生物反应器本身的面积为21,000cm2,相当于120个T-175培养瓶。值得注意的是,在细胞接种前,该生物反应器的中空纤维必须涂有粘合基材,如纤连蛋白、波形蛋白或冷沉淀物(通常来自多个供体的血液)。迄今为止,已有多达25项实验研究测试了Quantum®细胞扩增系统用于成人MSC扩增,使其成为贴壁细胞使用最广泛的生物反应器系统。此外,在比较脂肪组织来源的MSC(AT-MSC)的产量和群体倍增时,Quantum®优于基于培养瓶的手动培养,因此适用于大规模生产。此外,用人血小板裂解物(hPL)取代胎牛血清(FBS)作为生长补充剂,可显著增强AT-MSC在Quantum®内的扩增,同时保持其质量。例如,以20×106的密度加载到Quantum®中,来自骨髓的解冻MSC(BM-MSC)(在第2代)(在第2代)扩增7天,产量为100–276×106。此外,几项研究表明,能够在体外抑制T淋巴细胞活化的量子®扩增BM-MSC可以保持免疫调节功能。此外,Quantum®系统可以直接连接到任何气体及其组合,从而提供常氧或低氧微环境。Hanley等人观察到,与Quantum®和缺氧扩增的BM-MSC相比,培养瓶和常氧培养的BM-MSC的生产率较低。另一方面,加强对乳酸和葡萄糖水平的监测,再加上Quantum®生物反应器内新鲜培养基的持续供应,很可能促进了卓越的存活率和整体生长。通常,与基于培养瓶的增殖相比,Quantum®减少了所需的传代次数(减少到一半)和开放作(从54400个步骤减少到133个步骤)。此外,他们的研究还声称量子®扩增BM-MSCs在缺血性卒中大鼠模型中的治疗效果。另一项研究揭示了源自脐带组织(UC-MSC)的Quantum®制造的MSC和移植到诱导膝关节关节表面缺损小鼠身上的BM-MSC的愈合能力。采用Quantum®生物反应器系统的临床试验探索了GVHD、2型糖尿病、帕金森病、高级别神经胶质瘤、缺血性心脏病、中风和急性呼吸窘迫综合征等疾病的治疗方法。利用各种MSC类型,包括BM-MSC、AT-MSC或神经MSC,这些试验证明了使用Quantum®系统在不同剂量和给药途径下产生的细胞的安全性、耐受性和有效性。 来自MiltenyiBiotec的CliniMACSProdigy®设备利用ACC(贴壁细胞培养)工艺和适当的管路组(TS730),可实现自动MSC分离(通过骨髓样本的密度梯度离心)、接种、培养、培养基更换、繁殖和收获。Godthardt等人展示了该系统的大规模和临床级效率以及特定MSC-BrewGMP培养基的使用。此外,他们声称原代组织分离的BM-MSC以及AT-MSC或UC-MSC的单细胞悬液可以由CliniMACSProdigy®处理。此外,据报道,使用1层CellSTACK®的10天程序能够产生100多个集落,由29至5000万个MSC(第零代–P0)组成,这些集落是从马电针动员的外周血样本中分离出来的。分离的MSC具有特征性的成纤维细胞样形态和MSC表型特征。重要的是,与手动方案相比,Prodigy系统在P0收获的MSC数量显著更高,并且通过随后的手动传代提高了纯度,P3中CD73、CD90和CD105的阳性率达到95%。在制造商研究中,人骨髓单核细胞最初接种在1层CellSTACK®腔室中。然后在CliniMACSProdigy®系统内使用MSC-BrewGMP培养基洗涤、培养和扩增这些细胞,并将其扩增成三个5层CellSTACK®腔室。为了进行比较,使用T175培养瓶进行手动扩增过程。扩增14天后,CliniMACSProdigy®系统产生3.8×10⁸个BM-MSC(第2代),而手动过程在同一传代下产生4.5×10⁸个细胞,表明自动和手动方法之间的细胞数量相似。然而,平台复杂性和高耗材价格使CliniMACSProdigy®成为昂贵的制造选择,尤其是对于小规模MSC增殖。 NANT001或XL系统提供了另一个平台,在XL系统的情况下,包含一个CellSTACK®(636cm2)或两个互连的细胞培养容器(T-175和3180cm2的5层培养瓶)。与CliniMACSProdigy®不同,该系统不需要外部培养箱。另一方面,它的产能较低,但足以满足中小批量的自体生产。此外,它还允许对过程控制信息(例如温度、pH值、汇合度和细胞分离)进行智能和远程监控。NANTCartridge是一个封闭系统,由四个通过无菌连接器易于连接的部分组成[23]。Fitzgerald等人测试了NANT001系统在符合GMP的要求下生产来自健康供体的自体AT-MSC的效率。手动和自动过程同时进行,将加工后的基质血管组分(SVF)的细胞以4,000个细胞/cm2的密度放入一个636cm2的CellSTACK®培养室中。总体积为150mL的培养基由补充有5%hPL和1U/mL肝素的最低必需培养基(α-MEM)的α改良型组成。用150mL磷酸盐缓冲盐水洗涤培养样品,然后在第1天、第3天和50%汇合时更换培养基。在达到90%汇合度24小时后,用50mL重组TrypLE溶液收获扩增的细胞。来自三次验证运行的数据显示,分别持续6-8天和7-9天的手动和自动过程的细胞产量(平均2.7×107)和活力(>90%)相当。此外,两种扩增的AT-MSC都具有MSC表型特征以及与脂肪细胞和骨细胞相似的分化能力。 CellQualia™代表了另一个由Sinfonia技术设计的智能细胞处理系统。该系统使用两个细胞培养单元以及从1层到5层(CellSTACK)®和36层(HYPERStack®)腔室的自动传代,实现MSC的紧密和自动化生产。分析技术的内置过程通过对自动采集的样品进行在线(温度、CO2、形态、pH、葡萄糖和乳酸)和离线分析,确保高水平的质量控制。通过监测乳酸水平随时间的变化,可以间接评估细胞生长和增殖动力学。一旦乳酸水平达到表明最佳细胞密度的某个阈值,系统就会表明需要自动传代。此外,犬尿氨酸的积累和低水平2-氨基己二酸的维持表明细胞在整个过程中保持未分化状态。Hosoya等人证实了CellQualia™连续繁殖的MSC的塑料粘附和表型特征,这些MSC对MSC特异性标志物CD105、CD73和CD90呈阳性。然而,该研究缺乏对MSCs阴性标志物及其分化能力的评估。从2×10开始,该公司表示,在培养2次传代和6天后,MSC产量相当于1×108个细胞。 Lonza推出的Cocoon®平台旨在满足自体(患者规模)细胞疗法的生产要求。该系统使用封闭的一次性盒,为培养锚依赖性或独立细胞提供平面或体积空间。在包埋盒内,热障将温暖和寒冷区域分开,确保细胞培养(37°C)和试剂储存(4°C)的最佳条件。此外,该盒还配备了pH和溶解氧传感器,可在制造过程中实时记录数据。该平台是可定制的,易于使用,并且需要最少的作员干预,但是,目前其广泛的实施尚不清楚。 由Cytiva开发的Xuri™细胞扩增系统W25代表了细胞治疗生物工艺的飞跃。该系统采用单向WAVE™摇摆技术和各种培养体积(从300mL到25L),适用于小规模研究或大规模临床生产。WAVE™技术提供的轻柔摇动运动增强了营养物质的分布和气体交换,这是维持最佳细胞生长和活力的关键因素。专为免疫治疗中不依赖贴壁的细胞(嵌合抗原受体T细胞)的可扩展扩增而设计,但也可以参与微载体上的MSC生产。Silva等人在小体积接种到2L细胞袋的CultiSpher-S微载体上进行了UC-MSC的扩增,并在最初的24小时内保持静止。之后,在600mL培养基中的摇摆培养物中扩增UC-MSC。总体而言,该系统产生的膨胀系数范围为12.3至24.8。重要的是,摇摆培养中采用的动态条件不会对扩增细胞的质量产生负面影响,这通过各种质量控制措施(包括活力、表型保留和分化潜力)得到验证。有趣的是,另一项研究发现,在摇摆生物反应器中自发产生的MSCs聚集体表现出增加的干性、迁移特性和血管生成因子表达,从而提供了更高的治疗潜力。总体而言,Xuri™细胞扩增系统W25是一种多功能且高效的MSC扩增工具,为研究和临床应用提供了可靠的解决方案。3.细胞培养容器的放大 多层培养瓶是大规模MSC生产的重要组成部分,与传统使用的T-25、T-75和T-175培养瓶相比,其表面积更大(表1),从而减少了MSC的广泛传代培养。通常,放大涉及使用更大的生物反应器来提高生产能力,需要进行大量优化以保持更大体积的细胞质量。相比之下,横向扩展通过添加更多相同大小的生物反应器来扩大容量,确保各单元的条件一致,但需要更多的空间和资源。在放大和横向扩展之间进行选择取决于生产目标、预算和保持细胞一致性的能力等因素。如今,多层培养瓶是最有效的大规模策略之一,据报道其扩增率为100倍甚至更高。另一方面,随着细胞培养过程扩大到更大的容器尺寸,在整个生产过程中保持无菌和均一的培养环境变得越来越具有挑战性。表1.Corning和ThermoFisher不同多层培养瓶的生长面积 CellSTACK®多层培养瓶(Corning)和Nunc™CellFactory™系统(ThermoFisher)就是此类容器的示例,它们由堆叠在一起的多层细胞培养表面组成。它们都有5种尺寸,从1到40层不等,每层提供636厘米和632厘米的生长面积2。据报道,对于2室CellSTACK®,在5天的培养过程中,最终细胞产量为2.84×108(WJ-MSCs(沃顿氏果冻-MSCs)和4.69–5.65×107(BM-MSCs)。比较这两项独立研究,初始接种的细胞数量存在显着差异,范围为1000至4000个细胞/cm2,导致计算的扩增率存在195.28倍和9.22-11.10倍的巨大差异。同样,两位独立作者使用4室Nunc™CellFactory™在收获时获得了不同的细胞产量,对应于扩增的BM-MSC×7.8×108和2.5108。最后,在5腔CellSTACK®中培养牙髓来源的MSC(DP-MSC)11天,分别产生3.2×108个细胞在补充FBS和hPL的培养基中培养的108个细胞。总体而言,接种密度、培养基、某些添加剂和培养时间等细胞培养参数会影响给定系统的细胞产量。此外,康宁的CellBIND®表面处理通过增加聚苯乙烯细胞培养容器的亲水性来增强细胞粘附和生长,使其更有利于多种细胞类型。这种处理确保了一致、稳健的细胞培养条件,这对无血清或低血清培养基应用特别有益。 HYPERFlask®和HYPERStack®容器采用独特的细胞-介质环境气体交换技术,通过透气材料消除容器内的内部顶部空间。在补充有15%人AB血清的α-MEM中,以2×103个细胞/cm2的浓度接种10层HYPERFlask®(1720cm2)(共3.44×106个细胞)中,从人脐带基质(UCM-MSCs)衍生的扩增MSC能够在培养11天后产生4.5×107个细胞,相当于12.99的扩增率。HYPERStack®由Stackettes组成–一个单独的细胞培养室,由顶板和透气膜组成。每个Stackette拥有500cm2的生长面积,它们通过两个(液体和空气)歧管互连,形成一个模块,每个Stackette之间有一个露天气管空间。因此,HYPERStack®的12层和36层模块(相当于6000和18000cm2)具有与传统的2层和10层堆叠培养容器相同的体积占地面积。从而最大限度地减少培养空间,同时满足培养细胞的代谢需求。Sherman等人进行的初步数据显示,以3×103个细胞/cm2的密度接种BM-MSCs扩增5天后,可以®产生8.7亿个细胞的平均收获产量(扩增比为16.11)。 康宁®CellCube®系统为贴壁细胞培养提供了一个新颖、稳定且可扩展的平台。其模块化设计由平行的10、25或100个聚苯乙烯板组成,可在紧凑的占地面积中提供8500至85000cm2的大生长表面积。在CellCube®系统内,在每个培养板的两侧接种和培养细胞,使培养空间加倍。接种过程需要优化贴壁时间和系统的多次垂直旋转,以均匀接种细胞。此外,内置蠕动泵和一次性生物反应器可确保高效的连续流体再循环,以实现最佳的气体和营养物质输送。CellCube®系统的这种基于灌注的设计创造了一个与体内条件非常相似的环境,从而提高了细胞生产率。提供的信息概述了HEK293T和Vero细胞培养物在100层CellCube®模块中的成功扩增,展示了对培养条件的严格控制以及使用代谢物分析来确定收获时间。HEK293T和Vero细胞培养物均能有效扩增,分别×10个10个细胞的产量达到1.4和1.1。4.优化MSC生产:利用符合GMP要求的血清和无血清培养基 工艺验证是药物开发中的关键步骤,在此期间,必须仔细评估可能危及产品安全性的培养基、添加剂和材料的使用。例如,使用动物源性添加剂存在病原体传播的风险,需要进行彻底的风险分析并考虑替代添加剂。选择合适的培养基对于MSC的扩增和质量是必不可少的,尤其是在临床目的的大规模生产中。FBS作为富含生长因子和营养物质的天然补充剂,在许多实验室应用中用于细胞体外扩增,有着悠久的历史。然而,使用FBS培养旨在进一步用于治疗应用的MSC会带来重大风险,这主要是由于牛病毒、朊病毒和人畜共患病原体的潜在污染,这些物质可能会引发人类的免疫反应。在GMP级MSC生产中,利用经验证的FBS彻底检查免疫调节剂的存在,对于维持质量标准至关重要。γ辐照处理被认为是防止FBS中病毒污染的最佳方法之一,它与GMP流程一致。然而,采用无异种成分(xeno)的培养基,如富含人源物质的含血清培养基或无血清和无异种成分的培养基(SXFM),是符合GMP要求的MSC生产的最佳选择。将人类成分而不是动物衍生物加入培养基中,可确保符合细胞扩增过程的GMP指南。目前,包括人血清、脐带血血清、富血小板血浆和hPL的人源化补充剂被用作FBS的替代品(表2)。尽管自体人血清是MSC扩增的最佳选择,但获得足以产生临床相关量MSC的血清将具有挑战性。然而,来自多个供体的混合同种异体人血清可能有助于克服这一限制并促进MSC的大规模生产。尽管如此,人源补充剂(例如自体人血清)还有下一个显着的缺点,即从一个批次到另一个批次观察到的显着差异。尽管这些变化可以通过每批生产大型供体库来减轻,但它们同时也增加了可能因人类病毒而引起的感染风险。因此,这些障碍可能对大规模临床试验中MSCs的充分扩增和相关治疗效果构成重大挑战。 为了在不使用任何动物产品的情况下获得安全的人类产品,研究人员使用psoralen或核黄素灭活的hPL作为FBS的替代品,验证了在GMP条件下生产MSC。这种方法保留了MSC的多能和免疫调节能力。Mareschi等人使用hPL而不是FBS从5个骨髓样本中分离MSC。他们研究了一种新的hPL生产方法,该方法涉及用Ca-Gluconate处理血小板池以形成凝胶凝块,然后机械挤压以释放生长因子。将通过冻融循环和添加肝素获得的标准hPL(hPL-E)与新的hPL(hPL-S)没有血小板或纤维蛋白原,但含有相似数量的蛋白质和生长因子。hPL-S需要更少的生产步骤来符合GMP条件,保持干细胞标志物,并显示出显着差异,例如HLA-DR表达较低,使其成为MSC生产的有效替代方案。目前,有几种市售的病毒灭活GMPhPL以及最终的商业产品,如人血小板裂解物(生命科学生产)、血小板裂解物MultiPLi(Macopharma)和PLUS™GMP级(CompassBiomedicalInc)。此外,与hPL相关的污染风险得到有效管理。例如,自2018年以来,法国的所有血小板浓缩物都经过补骨脂素处理,以降低微生物污染的风险。另一种消除潜在病原体的方法是在使用hPL之前对其进行伽马射线照射[57]。然而,与hPL产品生产相关的一个重大限制是必须确保血小板供体的充足供应。管理符合GMP的培养基的补充选择是使用合成的、化学成分明确的培养基,例如BDMosaic™间充质干细胞无血清(BDBiosciences)、MesenCult-XF™(StemCellTechnologies)、MSCNutriStem®XF(生物工业)、无血清成分确定的细胞培养物(TNCBioBV)等。上述培养基的成分仅由已知化合物组成,使其成为FBS的潜在可取替代品。尽管如此,它们特别适用于研究目的,因为它们需要不断开发和改进以维持关键的细胞功能,例如粘附、分化、免疫调节和其他基本过程。用于优化和可重复的MSC生产的下一组培养基是具有高质量成分的无血清、无异种成分配方,例如MSC-BrewGMP培养基(MiltenyiBiotec)和StemProMSCSFMXenoFree™(Invitrogen)等。然而,化学成分明确的培养基的一个关键限制是由于缺乏附着因子,MSC增殖缓慢或有限,以及细胞对底物的粘附不足。因此,使用某些培养基需要在塑料表面预涂特定物质,以确保最佳的细胞附着和生长。另一方面,补充人血浆衍生物的StemProMSCSFM™(LifeTechnologies)等培养基对于无包被的MSC培养来说并不是最佳。MSC-BrewGMP培养基(MiltenyiBiotec)是市场上唯一一种符合GMP要求的SXFM,因为它是袋装的,用于封闭系统培养,不需要表面涂层。最常用的分子,如纤连蛋白、层粘连蛋白和肽,用于表面改性,通过引入正表面电荷来促进细胞生长,从而增强粘附力。表2:用于 GMP 级 MSC 生产的培养基 比较补充动物或人血清的不同培养基类型与无血清培养基(SFM)的效果,可得出对MSC特性的显著结果。Dreher等人证明,与在10%FBS中培养的AT-MSC相比,在补充有10%人血清的培养基中培养的AT-MSC在应用后在非肥胖糖尿病免疫缺陷小鼠肺和肝脏中的存在减少。总之,在人血清存在下培养的AT-MSC在特异性粘附和细胞外基质相关分子的下调方面表现出有趣的变化。他们认为整合素α6(CD49f)表达的降低可能与对层粘连蛋白的粘附减少有关。尽管如此,减少间充质干细胞在肺部的滞留可能会降低肺栓塞的风险。细胞扩增培养基的选择在影响MSC的增殖动力学等特性方面起着关键作用。多项实验研究揭示了各种培养基类型对MSC特性和培养动力学的显着影响。Wuchter等人进行了一项比较分析,研究了三种符合GMP的培养基对MSC扩增的影响:补充有10%混合hPL的基础培养基(Dulbecco改良Eagle培养基(DMEM)-低葡萄糖)、StemPro®MSCSFMCTS(Invitrogen)(用于人离体组织和细胞培养加工应用)和补充有10%FBS的非XFMSCGM™(Lonza)(仅供研究应用)。最显着的差异出现在延长培养(50天)后BM-MSCs的大小和增殖动力学。StemPro®MSCSFMCTS显著促进了生长,与其他MSC相比,产生的MSC具有更小但高度均匀的形态。在hPL培养基中培养的BM-MSCs表现出明显的突起,但与在含有FCS的MSCGM™中培养的BM-MSC相比更小。然而,与在MSCGM™中培养相比,用hPL培养基培养的MSC观察到的生长增加略大。血小板裂解物对MSCs扩增的促进也与其他研究的结果一致。此外,Poloni等人证明,在含有10%混合同种异体人血清的Iscove改良Dulbecco培养基中培养的CD271阳性BM-MSC与在补充动物血清的培养基中培养的培养基相比,表现出相当的分化能力以及表面和基因标志物的表达。此外,未观察到核型改变。他们还证明,使用CD271选择的细胞和混合的同种异体人血清,仅30天内从5mLBM抽吸物中细胞产生显着增加到临床显着水平(10×10^8MSC)[66]。此外,SFM的使用也对MSC的特性产生了显着影响。例如,PRIME-XVSFM(IrvineScientific)已被证明可以增强人BM-MSC中的集落形成能力,提高成骨潜力,并增加群体倍增。因此,它是动物血清的优质替代品。尽管基于FBS的培养基和PRIME-XVSFM的渗透压相似(分别为0.31和0.29Osmol/kg),但SFM更接近人类生理条件,因此被认为对细胞有更好的影响。此外,Aussel等人证明,SXFM,特别是MSC-BrewGMP培养基(MiltenyiBiotec),显著影响MSC的克隆形成效率。具体来说,与传统的AT-MSC和BM-MSChPL培养基相比,SXFM条件下的菌落大小和密度增加。 值得注意的是,用于收获和分离用于临床应用的MSC的其他补充试剂也必须满足所有安全标准。目前,大多数临床研究使用猪来源的胰蛋白酶进行细胞分离制备。然而,为了优化该程序,使用TrypLE(Gibco,ThermoFisherScientific)或TrypZean(Sigma-Aldrich)等重组胰蛋白酶是有利的,因为它们对细胞的影响更温和,并且与猪来源的胰蛋白酶相比不存在异种蛋白。对于GMP条件下的酶组织消化,建议使用GMP级胶原酶而不是I型胶原酶,因为后者可能含有可能携带传染源的成分。出于这些目的,例如,源自溶组织梭菌的胶原酶NB6GMP级(NordmarkBiochemicals)是一种用于分离干细胞的高纯度组织解离酶。它包含具有酶活性的蛋白质,包括I类和II类胶原酶、中性蛋白酶和梭菌蛋白酶。0.4PZU/mL胶原酶NB6的最佳浓度和3h的消化时间表明,在第0代时,沃顿胶溶胶衍生的MSC产量更高。此外,其他类型的胶原酶也用于组织消化:CLSAFA(Worthington)和LiberaseMTF-SGMPGRADE(Roche)。另一方面,胰蛋白酶和胶原酶的另一种替代替代品是Accutase(Life Technologies),它是蛋白水解酶和胶原水解酶的混合物。然而,离心、过滤或微碎裂等机械作提供了避免与胶原酶使用相关的安全问题的替代方法。根据迄今为止收集的数据,来自骨髓、脂肪组织和脐带的MSC是临床试验中最常用的细胞来源。然而,AT-MSC在大规模生产中具有优势,因为与来自同一供体的BM-MSC相比,AT-MSC在长时间培养过程中具有增强的遗传和形态稳定性,并且增殖更快。此外,AT-MSCs在25次传代后表现出分化潜能降低,而BM-MSCs的免疫抑制作用仅在7次传代后观察到下降。5.临床条件下MSC的冷冻保存 细胞或组织在细胞疗法中的应用导致了治疗获得性或遗传性疾病的新型药物的诞生。这些是已知的ATMP,被定义为主要生物作用由细胞或组织执行的治疗产品。冻存保存仍然是延长细胞(包括MSC)保存时间的唯一技术。它有助于维持细胞功能特性,并允许细胞聚集以达到临床使用所需的数量。与成骨细胞和软骨细胞等特殊细胞的冷冻保存相比,MSC必须考虑更多因素。具体来说,评估MSC冷冻保存后的免疫调节和多系分化能力、细胞活力和表型至关重要。如果这些能力受损,MSC可能无法保持原有的免疫调节和再生特性。冷冻保护剂 冷冻保护剂是能够防止生物物体遭受冰冻损伤并确保其解冻后存活的试剂。冷冻保护剂在冷冻过程中的保护作用基于它们与细胞内外的水分子形成牢固键的能力,这比水分子本身之间的键更强。此外,它们还可以降低盐浓度,最大限度地降低破坏细胞蛋白质结构的风险。低温保护剂还与膜的结构部件粘合,保护它们免受冰晶的损坏(图2)。图2:细胞冻存机制作为基于细胞的疗法生产的关键步骤。在冷冻保存过程中导致细胞损伤的过程涉及特定的机制。为了防止这种伤害,必须在细胞悬液中添加适量的适当冷冻保护剂(CPA)并应用最佳冷却速率。 已经开发了各种技术来冷冻保存MSC,例如慢速冷冻和玻璃化。但是,这些方法有局限性。首先,即使使用冷冻保护剂(CPA),使用这些方法保存的MSC仍然存在冷冻损伤的风险。其次,二甲基亚砜(DMSO)等CPA可能会无意中诱导MSC分化为神经元样细胞。为了解决这些问题,需要优化CPA的使用或开发新的冻存方法。微/纳米技术的最新进展提供了有希望的改进,有可能提高冷冻保存的效率并克服当前方法的局限性。在高度疏水性的纳米粗糙表面上冷冻封装在纳升液滴中的细胞以及创建无毒的纳米级生物启发CPA等创新是该领域特别有前途的进展。 已建立的MSC冻存方案使用DMSO。残留的痕量DMSO,即使在洗涤步骤之后,也会对患者造成重大不良反应的风险。此外,DMSO与异常基因表达和MSC分化有关。因此,开发一种不含DMSO的MSC冻存方法是非常可取的。海藻糖、植物蛋白和防冻蛋白模拟物等物质已与降低浓度的DMSO相结合,以改善MSC冻存。这些组合成功地抑制了冰的再结晶,并保护了细胞膜,类似于天然防冻蛋白。目前有几种市售的冷冻保存选项用于MSC的冷冻保存。 CryoStor®CS2、CS5和CS10是市售的GMP级冻存培养基,设计用于优化细胞和组织在−80°C至−196°C的超低温环境中的保存。这些制剂在冷冻、储存和解冻过程中提供保护环境,增强细胞活力和功能,同时无需血清和高细胞毒性试剂。CryoStor专门处理细胞的分子生物学方面,减少冻存诱导的延迟性细胞死亡,从而提高解冻后活力,特别是对于MSC和肝细胞等敏感细胞类型。该产品线包括CryoStorCS2(2%DMSO)、CS5(5%DMSO)和CS10(10%DMSO)。冻存过程涉及关键步骤。最初,通过机械或酶解离制备细胞,然后离心以产生细胞沉淀。去除上清液,同时尽量减少培养基稀释。然后加入CryoStor冷培养基,使其浓度达到0.5-10×10^6个细胞/ml,在2-8°C下孵育约10分钟。以每分钟-1°C的受控速率将温度降低到-80°C,在此温度下保持15-20分钟,然后在-5°C左右开始冰成核,从而开始成核。或者,样品可以在−20°C下保存2小时,然后在−80°C下保存。冷冻后,它们可以在低于-130°C的液氮中储存,或在-80°C的温度下短期储存。解冻至关重要,应在37°C水浴中迅速解冻。解冻后,细胞-CryoStor混合物需要用培养基在20–37°C下稀释。解冻后活力应在24小时后使用活/死荧光测定或MTT等代谢测定进行评估,以仔细比较与非冷冻对照的准确性。 Ho等人提出了一项研究,其中犬AT-MSC被修饰为过表达治疗性转基因,用CryoStor10冷冻保存长达11个月。结果表明,冻存不会影响转基因表达、细胞活力或迁移能力。值得注意的是,解冻的MSC表现出与新鲜MSC相当的细胞毒性,可有效治疗患有癌症的犬患者,并导致超过20个月的无进展生存期,突出了冻存MSC在临床应用中的可行性。此外,将马MSC冷冻保存在CryoStor®CS10中,并评估其在小马体内的活力。Williams等人的研究涉及在模拟冷藏运输后和解冻后立即通过静脉注射这些MSC,并监测临床和血液学变化。结果显示没有不良反应或血液参数偏差。注射后CD4+和CD8+淋巴细胞群增加,表明可能存在免疫反应。CS10有效地维持了MSCs的活力。有必要进一步研究以探索与不同MSCs供体的免疫相互作用。 最近的一篇论文比较了两种冷冻保护剂:STEM-CELLBANKER™(CB)和10%DMSO,均在无异种成分培养基中,用于AT-MSC和BM-MSC。与非冻存MSC相比,两种冻存保护剂都保持了相似的细胞形态和表面标志物表达。然而,冷冻保存的AT-MSC与CB(90.4%)相比于DMSO(79.9%)具有更好的活力。虽然在CB和非冻存条件下,AT-MSC的群体倍增时间相当,但使用DMSO时略有增加。BM-MSC显示两种冷冻保护剂的倍增时间更长。重要的是,两种MSC类型在冷冻保存后都保留了其分化能力。总之,CB比DMSO更能保持AT-MSC的特性,这可能会影响细胞治疗应用。据制造商称,STEM-CELLBANKER®DMSOFREEGMP级是一种创新的冻存解决方案,旨在保存不含DMSO的各种细胞类型。其配方符合JPN、EU、US和PIC/S的GMP指南生产,不含血清、动物源性成分和DMSO,减轻了对研究至关重要的纯度的担忧。STEM-CELLBANKER®DMSOFree可显著提高细胞活力,同时保留细胞多能性和解冻后有效增殖等关键特性。该解决方案提供了卓越的批次间稳定性,确保了批次间性能的一致性。制造商还声称用户会喜欢它的便利性,因为它不需要准备;细胞可以直接在-80°C下冷冻,从而简化了冻存工作流程。它还在美国FDA药物主文件中注册,允许客户申请监管提交授权书。总体而言,该产品代表了那些寻求无DMSO解决方案的人在低温保存方面的重大进步。 人脐带(UC)是MSC的一个有前途的来源,尽管被归类为实体组织,但UC具有单个供体的多种用途等优势。然而,以前使用动物或同种异体血清的冻存方法导致冻融UC-MSC中的细胞增殖较低。Shimazu的研究团队使用无血清和无异种成分的冷冻保护剂STEM-CELLBANKER建立了一种最佳的冷冻保存技术。经过缓慢的冷冻过程和随后的解冻,保存的UC产生的MSC保持了与新鲜细胞相当的典型表型、免疫抑制能力和分化潜力,证明了临床活力。冻存方法和可用系统 冻存MSC有两种主要方法:慢速冷冻和快速冷冻/玻璃化。缓慢冷冻的速率为1°C/min,因其污染风险低且更易于加工而受到诊所和研究实验室的青睐,允许在一个含有低浓度CPA的小瓶中冷冻大量MSC。尽管存在冻伤风险,但优化CPA的使用对于防止冰晶形成至关重要。玻璃化反应涉及使用液氮将细胞悬液从水相快速转化为玻璃态,需要多摩尔CPA混合物和逐步引入,以尽量减少化学毒性。然而,它有渗透损伤的风险,并且由于需要人工作和高技能,不太适合大体积处理。 开发了一种新的低温保存方案,使用甜菜碱和电穿孔。甜菜碱是一种天然的渗透保护剂,稳定、无毒且高度亲水性,这使其能够竞争性地结合水分子并防止结冰。电穿孔被广泛接受用于将非渗透性分子输送到细胞中,在电刺激下在细胞膜上产生临时的疏水孔,允许分子通过。这种联合方法在各种MSC类型中表现出普遍适用性,并在各种MSC类型中保持较高的解冻后细胞活力,包括人源性UC-MSC、小鼠源性BM-MSC和转基因UC-MSC。 对于同种异体“现成”临床应用,需要大量的冷冻MSC。高细胞浓度的冻存可最大限度地减少DMSO输注量,从而降低输注相关毒性。使用CellSeal®冻存管,将1000万个/mL的细胞用5%DMSO冷冻至-80°C,并储存在液氮中。使用该系统冷冻的AT-MSC在解冻时显示出高功能活力。该系统可轻松进行床旁解冻和给药,残留DMSO极少,无需细胞洗涤。未来的研究将检查解冻后AT-MSC在体内的植入潜力。 在表3中,我们列出了用于MSC冻存的控速冷冻系统的市售选项。这些先进的冷冻系统在确保MSC在冷冻保存过程中的活力和功能方面发挥着至关重要的作用,允许受控的温度曲线,防止冰晶的形成并保持细胞完整性。该表提供了各种型号的详细信息,包括其规格、特点和典型应用,可帮助研究人员和从业人员在为其MSC冻存需求选择合适的设备时做出明智的决定。表3.用于MSC冻存的市售受控冷冻系统概述 关于MSCs悬液的冻存,必须专注于标准化细胞培养质量的评估。特别是,冷冻保存前后评估标准的一致性至关重要。通过比较冷冻前后的相似指标,我们可以准确确定冷冻保存对细胞培养的影响。此外,重要的是要认识到细胞培养物在冷冻保存后必须经过修复或驯化过程。几项研究表明,如果没有这个适应阶段,就无法完全恢复冷冻保存的干细胞的治疗潜力。 重要的是要认识到,各种复杂的基于细胞的产品可能有很大差异,这意味着单一的冻存方案可能并不适合所有产品。因此,为每个产品创建单独的协议会使标准化工作复杂化。因此,基于复杂生物医学细胞的产品的冷冻保存是一个具有挑战性和争议性的问题。然而,研究表明,这个概念是可以实现的,值得进行更深入的调查。阻碍冻存标准化的另一个方面是实验室之间技术设备的显著差异。通常,每个实验室都会根据其特定的技术能力和可用试剂来建立其冻存方案。因此,需要一种综合方法来标准化冷冻保存过程。尽管如此,编制通用指南以增强冷冻保存过程,最大限度地减少对细胞的压力应该是可行的。作者希望这篇综述有助于组织和选择冷冻保存MSCs、含MSCs的组织和相关生物医学细胞产品的方法。6.结论 当前用于生产临床级MSC的大规模扩增平台的一个局限性是不同系统和方法之间的细胞产量和质量的差异。虽然已经开发了各种生物反应器和培养容器来满足放大生产的需求,但在培养条件、细胞处理技术和质量控制措施方面仍然缺乏标准化。这种可变性会导致最终产品不一致,从而影响其治疗效果和安全性。此外,与其中一些平台相关的复杂性和成本可能会限制其广泛采用,尤其是在资源受限的环境中或小规模的生产需求中。 展望未来,应将工作重点放在标准化方案和优化大规模扩增平台上,以确保临床级MSC生产的可重复性、一致性和成本效益。这可能涉及开发更加用户友好的模块化系统,以便无缝集成到现有的制造过程中,以及实施先进的监测和控制技术,以确保对培养条件的精确调节。此外,需要继续研究新型培养基底物、培养基配方和生长因子,这些物质可以增强细胞增殖、维持干性并促进治疗效力。MSC的冷冻保存对于在临床应用中保持其活力和治疗潜力也至关重要。虽然DMSO通常用作冷冻保护剂,但它存在毒性和不必要的分化等风险,这促使开发了替代冷冻保存方法和试剂以及市售的GMP级冷冻保存培养基。此外,受控速率冷冻系统的进步提高了MSC解冻后的活力。涉及工业界、学术界和监管机构的合作倡议对于加速基于MSC的疗法从实验室到临床的转化至关重要。最终,应对这些挑战和提高大规模生产能力对于实现基于MSC的再生医学的全部潜力和满足对基于细胞的疗法日益增长的需求至关重要。识别微信二维码,添加抗体圈小编,符合条件者即可加入抗体圈微信群!请注明:姓名+研究方向!本公众号所有转载文章系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系(cbplib@163.com),我们将立即进行删除处理。所有文章仅代表作者观点,不代表本站立场。
细胞疗法免疫疗法临床研究
2025-04-19
·医药速览
概述我国类风湿性关节炎(RA)的患病率约为0.42%,患者总数约500万,男女比约为1∶4。RA的临床表现为滑膜炎,并逐渐出现关节软骨和骨破坏,最终导致关节畸形和功能丧失,可并发肺部疾病、心血管疾病、恶性肿瘤、骨折及抑郁症等。Update on the Pathomechanism, Diagnosis,and Treatment Options for Rheumatoid Arthritis,Cells 2020, 9, 880; doi:10.3390/cells9040880类风湿关节炎(RA)的免疫发病机制遗传因素100 多种等位基因多态性与疾病风险相关,MHC II 类等位基因尤其是HLA - DRB1等位基因关联性强。HLA - DRB1 等位基因含有在 β 链氨基酸残基 71 - 74 处的特定序列(“共享表位”),与氨基酸残基 11 和 13 共同形成抗原结合口袋,这种结构与类风湿因子(RF)和抗瓜氨酸化蛋白抗体(ACPA)的产生相关。T 细胞能够在 HLA - DRB1*04 的背景下识别瓜氨酸化抗原,而自身免疫性 B 细胞反应涵盖了大量瓜氨酸化蛋白质,这表明 MHC II 类分子在呈递抗原给 T 细胞,进而驱动 B 细胞产生自身抗体的过程中起到了关键作用,体现了 MHC II 类多态性与自身抗体产生之间的紧密联系。环境因素肠道菌群等环境暴露可能触发先天免疫,影响蛋白质抗原修饰,导致自身耐受性破坏。肠道菌群可以调节多种免疫细胞的功能,如调节 T 细胞(Treg)和辅助性 T 细胞(Th)的分化和活性。在 RA 患者中,肠道菌群的异常可能导致免疫细胞功能失调,例如使 T 细胞分化偏向促炎表型,从而促进炎症反应。正常情况下,肠道菌群有助于维持免疫系统的耐受状态。然而,在 RA 发病过程中,肠道菌群的改变可能破坏这种耐受平衡,引发自身免疫反应。例如,肠道菌群的失调可能影响肠道黏膜屏障功能,使抗原更容易进入体内,进而激活免疫系统,导致自身抗体的产生。鉴于肠道菌群在 RA 中的重要作用,调节肠道菌群可能成为一种新的治疗策略。例如,通过益生菌、益生元和粪菌移植等方法来改善肠道菌群的组成和功能,有望恢复免疫系统的平衡,减轻 RA 的炎症反应,为 RA 的治疗提供了新的方向。免疫细胞与信号网络异常多种免疫细胞类型和信号网络功能失常,引发适应不良的组织修复过程,主要损害关节,也可累及肺和血管系统。T 细胞免疫反应:HLA - DRB1 位点与 RA 相关,CD4⁺T 淋巴细胞驱动 RA,IL-6、IFN-γ、IL-17 等细胞因子参与,Th1、Th17 细胞起重要作用,T 细胞通过多种机制迁移至关节,其可塑性对免疫应答有重要意义,但功能作用尚未完全明确,调节性 T 细胞(Treg)功能可能受损,细胞因子 IL - 22 也参与炎症反应。B 细胞免疫反应自身抗体产生:自身抗体主要由 B 细胞产生,RF 和 ACPA 是重要的 RA 相关自身抗体,ACPA 对早期诊断和预测 RA 特异性高,其可通过多种机制诱导炎症,如激活免疫细胞、诱导 NETosis、激活补体系统、促进破骨细胞分化等,RF 在诊断中有一定价值但水平与疾病活动相关性存在争议。抗原呈递:B 细胞是重要抗原呈递细胞,在 RA 中可向 CD4⁺T 辅助细胞呈递自身抗原,循环中 T 滤泡辅助细胞(Tfh)和外周辅助细胞(Tph)频率增加,Tph 细胞可促进 B 细胞活化、增殖、分化和抗体产生,血清 IL - 21 水平与 RA 疾病活动相关指标呈正相关。细胞因子分泌:RA 患者外周血 B 细胞分泌多种细胞因子,TNF - α 可促进破骨细胞形成,调节性 B 细胞(Breg)分泌抗炎细胞因子抑制 RA 进展,活动期 RA 患者外周血中具有调节功能的 B 细胞数量减少。破骨细胞激活:记忆 B 细胞表达 RANKL,调节骨稳态,ACPA 阳性 RA 患者骨吸收更明显,血浆细胞也可促进破骨细胞生成。先天免疫介导的免疫反应:先天免疫系统在 RA 起始和进展中起重要作用,巨噬细胞是 RA 滑膜中最丰富的免疫细胞,产生多种炎性介质,M1/M2 巨噬细胞失衡与 RA 相关;树突状细胞(DC)在诱导耐受或自身免疫中起作用,其表型和功能复杂;自然杀伤(NK)细胞和先天淋巴细胞(ILC)在 RA 发病中也有一定作用,ILC 分布变化与 RA 发展相关,IL - 9 产生的 ILC2 细胞可促进炎症消退。Jang, S.; Kwon, E.-J.; Lee, J.J. Rheumatoid Arthritis: Pathogenic Roles of Diverse Immune Cells. Int. J. Mol. Sci. 2022, 23, 905. https:// doi.org/10.3390/ijms23020905不同阶段的检查点变化检查点 1:全身性自我耐受崩溃初始耐受破坏位置:关节外,如流行病学研究发现 RF 在滑膜炎前多年存在,抗瓜氨酸化和氨甲酰化抗原的自身抗体也在临床前期出现。免疫应答特点:RA 自身免疫针对翻译后修饰蛋白,具有高度多反应性,主要由外周耐受缺陷主导,遗传风险因素促进自身抗体产生,T 细胞在自身抗体产生和疾病进展中起关键作用。检查点 2:从无症状自身免疫到组织炎症的转变炎症触发因素:环境暴露等因素使疾病过程转移,免疫细胞进入滑膜,肠道微生物群等环境因素可能是风险因素,早期滑膜炎病例中 CD3⁺T 细胞常见,滑膜活检样本组织学表型可预测疾病。T 细胞异常与功能后果:CD4⁺T 细胞内在缺陷导致分化异常,与 DNA 修复机制缺陷和细胞生物能量重编程有关,如 ATM 和 MRE11A 表达降低影响 DNA 修复,导致端粒脆弱、线粒体 DNA 损伤等,使 T 细胞分化为短寿命效应 T 细胞(SLEC),引发滑膜炎症,还可能导致 T 细胞记忆反应减弱、寿命缩短、增殖压力增加等,影响宿主免疫力,如患者中 CD4⁺CD28⁻T 细胞具有早衰特征,与冠状动脉疾病存在共同发病机制。检查点 3:急性滑膜炎向慢性持续性滑膜炎的转变组织细胞变化:慢性滑膜炎使免疫细胞增殖压力增加,部分患者病情进展,其发病与局部组织环境相关,如滑膜成纤维细胞活化、组织侵袭性 T 细胞诱导炎症和淋巴样结构形成等,应用高分辨率技术发现滑膜组织中存在多种细胞亚群,包括成纤维细胞、巨噬细胞、T 细胞和 B 细胞等,各细胞亚群具有不同特征和功能,如部分成纤维细胞为促炎效应细胞,巨噬细胞有保护性和促炎性不同亚群,T 细胞包括多种功能亚群,B 细胞在滑膜炎症中也有一定作用,且滑膜 T 细胞和巨噬细胞是 TNF 的细胞来源,TNF 在 RA 滑膜炎中起重要作用。推文用于传递知识,如因版权等有疑问,请于本文刊发30日内联系医药速览。原创内容未经授权,禁止转载至其他平台。有问题可发邮件至yong_wang@pku.edu.cn获取更多信息。©2021 医药速览 保留所有权利往期链接“小小疫苗”养成记 | 医药公司管线盘点 人人学懂免疫学| 人人学懂免疫学(语音版)综述文章解读 | 文献略读 | 医学科普|医药前沿笔记PROTAC技术| 抗体药物| 抗体药物偶联-ADC核酸疫苗 | CAR技术| 化学生物学温馨提示医药速览公众号目前已经有近12个交流群(好学,有趣且奔波于医药圈人才聚集于此)。进群加作者微信(yiyaoxueshu666)或者扫描公众号二维码添加作者,备注“姓名/昵称-企业/高校-具体研究领域/专业”,此群仅为科研交流群,非诚勿扰。简单操作即可星标⭐️医药速览,第一时间收到我们的推送①点击标题下方“医药速览” ②至右上角“...” ③点击“设为星标
临床研究免疫疗法微生物疗法
2025-04-15
·动脉网
成为一名免疫基因学者,获美国免疫学博士学位,被誉为“血清之父”,成为著名的骨髓移植配型、干细胞临床实验室专家,任美国华盛顿大学教授、美国红十字总会研究所HLA实验室主任、世界红十字会第一届世界组织相容性会议主席,慈济骨髓捐献资料中心主任等职务,并于1992年回到中国,致力于建立华裔骨髓资料库。以上似乎是李政道博士为人津津乐道的前半生的总结。这样的经历背后,承载的是20世纪下半叶免疫学和再生医学飞速发展的历程,自19世纪末期,俄国动物学教授Metchnikoff通过被刺伤的海星,看到炎症反应的第一步以来,免疫学认知在一个世纪内的时间里快速迭代,免疫学理论构架逐渐扩充、丰满,并在干细胞技术开启系统性研究后拓展版图,为尚未治疗的疾病和患者打开一扇新的大门。信仰“医学有着不争的普世精神——人的生命高于一切,生命面前人人平等”的李政道在大洋彼岸目睹了种种变革:基于干细胞的疗法,包括人类多能干细胞(hPSCs)和多能间充质干细胞(MSCs),已成为再生医学的关键参与者。美国癌症免疫疗法和欧洲间充质干细胞 (MSC) 疗法的成功批准使再生医学成为重要的治疗方式,且治疗方法侧重于疾病的管理而不仅仅是治疗。进入21世纪的第二个十年,李政道仍以90岁高龄坚持在科研前线。近日,动脉网专访了李政道博士,深入了解他与团队正在推动的再生医学项目群。这个旨在“让银发族活得有尊严”的项目,不仅将引入全球顶尖的质量标准,更包含多项突破性技术创新,有望成为中国再生医学发展的新引擎。01“血清干细胞之父”的基建工作李政道的学术生涯始于免疫学,并在这一领域取得了诸多开创性的成果。1968年,李政道在加拿大麦吉尔大学(McGill University)获得免疫学硕士学位,随后于1972年在美国罗格斯大学(Rutgers University)获得免疫学博士学位。而他的研究生涯从美国斯隆·凯特琳癌症中心(SKI)开始,在那里,他协助创建了骨髓移植项目,并在全球首次筛选到HLA-DR抗原。这一发现不仅丰富了人类基因库,还为器官移植、骨髓移植、干细胞移植、输血医学、亲子鉴定、法医学等领域带来了革命性的变化。在担任美国红十字会总会实验室主任及美国国立卫生研究院(NIH)血清库主任期间,李政道筛选并获得了全球最多的HLA血清抗体,占全球总量的约60%。他的研究成果不仅在学术界引起了广泛关注,还为全球的医疗实践提供了重要的技术支持。包括法国诺贝尔奖获得者的医生,也需要李政道团队提供整套的血清抗体,去分析病人的基因。此外,李政道还创建了DNA高解析方法,提供了解决组织相容性问题的第二类型的DR抗体。进一步推动了再生医学的发展,并进入全球顶级科学家的行列。值得一提的是,李政道也是全球最早将再生医学和计算机结合的科学家,在他的推动下,再生医学相关产品的出品周期缩减了90%,并能以竞争对手1/15价格出售。在此期间,一种科研成果转化的思路在李政道的学术和研究生涯中逐渐明晰。李政道曾坦言:“医学是我的信仰,但好的医学应该让更多人受益。科学家最大的幸福,就是看到自己的研究真正帮助到需要帮助的人。”这一理念,是一道指向未来漫长技术发展的射线,在免疫学理论、肿瘤治疗、再生医学技术迭代更新,甚至在这些技术不断迎来产业落地机会时,这种转化落地思维带来的影响都会被重新唤起。这便是行业基建的力量。中华医学会首次访问美时,李政道博士已在行业树立了威望,遂被邀请回国讲学一个月。之后李政道博士每年继续回国致力于免疫基因及干细胞的研究,并将其在中国发扬光大。1997年,回国后的李政道协助建立世界上第一个最大的华人骨髓库。他没有中断这些基建工作。李政道先后受邀在协和医科大学、华西医科大学、浙江大学等数十家知名大学院校担任名誉教授,并多次受到前国家国务委员李铁映、前卫生部部长陈敏章接见。值得一提的是,他捐赠给杭州的一辆无偿献血车,成为市内的第一辆无偿献血车,这是推进中国献血制度由有偿到无偿改革的一大步,提高了国内献血安全及输血安全,并推动中国血液界与国际接轨。与此同时,在学术和科研生涯中的“基建”经验也让他在中国的再生医学发展史上扮演了重要角色。他通过在国内担任多个顾问和名誉教授职位,无偿培养了大量再生医学人才。他还在国内无偿捐献了价值千万人民币的再生医学设备,推动了国内骨髓移植、器官移植以及干细胞事业的发展。2025年,李政道迈入上寿之年,他的脚步仍然没有停下。02再生医学项目群:让银发族活得有尊严“医学不应该只是延长寿命,更要让生命的每一刻都有尊严。”在问及为何要启动再生医学项目群时,李政道如是说。这或许正是这些年扎根免疫学、干细胞技术、肿瘤治疗的李政道在医疗健康观念的体现——从单纯治病转向全面提升生活质量。据悉,项目群将建设亚洲最严谨的再生医学临床实验室、引进被誉为行业质量金牌标准的AABB协会、应用李政道博士迭代50年的再生医学质量管理系统、应用AI程序对再生医学进行数据管理,上述项目类似于再生医学行业的基础设施,将保证提供“安全、有效”的产品,充分满足消费者对“安全、有效”的刚性需求。这也是李政道首次在大陆地区亲自操盘的再生医学项目群,愿景是通过本项目,打造再生医学行业的若干基础设施,服务于行业持续增长的各项需求,并“让银发族活得有尊严”。项目名称中的“银发族”一词,正体现了这种人文关怀。在人口老龄化加剧的今天,这种以提升老年人生活质量为目标的医疗创新,具有特殊的社会意义。■ 一根脐带服务1万人次项目群还将启动全球独创的技术,已经完成研发工作,并能够直接执行。这项技术的核心在于,可以让一根脐带服务1万人次以上。“传统方法培养的脐带组织最多治疗两个人。”李政道解释道,“而我们的技术可以让一根脐带培养出足够治疗上万人的细胞量。”具体到制备方式,前期工作包括将脐带切成若干小片,分别储存在1毫升的小试管里面,放置在液氮桶,需要的时候就取出一小瓶,可以扩增出上百亿上千亿的细胞,最多可以服务一万人次。由此一来,这项突破性技术解决了细胞治疗行业三大痛点:● 成本问题——原材料利用率提升上万倍,大幅降低治疗费用;● 来源问题——突破原材料稀缺限制,确保供给稳定;● 质量问题——标准化流程确保细胞质量一致性和安全性。据李政道介绍,这项技术培养到第三代时就能达到服务500人份的效果。随着研发工作的进行,当前该技术还实现了根据不同的种族将人群细分为20-30个族群,并按照不同族群,匹配相对应基因的干细胞产品。此外,这种方式能够避免排异反应,实现细胞产品产量扩大百倍。这意味着,未来再生医学的供给模式有望得到革新,同时治疗方式也将真正实现个性化定制,大幅提升治疗效果。■ “安全+有效”,撑起最严质控体系而在实现细胞产品有效扩增的背后,是严格的质控体系。“欧美的临床实验室管理非常严格。”李政道指出,“临床实验室的产品直接用于病人治疗,实验室主任通常是MD或PhD毕业,还要经过两年专业培训才能参加考试,录取率仅有10%。”值得一提的是,细胞及外泌体是药物等级的产品,其生产特点是“过程即产品”,因此对过程管理的要求极高,整体安全、全程不被污染成为该行业的核心竞争力。此外,对质控体系的严格把握,也和“让银发族活得有尊严”的理念息息相关。一根脐带服务1万人次的核心技术解决了成本、来源和质量问题,而要使其真正普惠大众、尤其高龄群体,还需要在产品落地环节精益求精。“质量安全是行业发展的基础,”李政道强调,“没有这个基础,再好的技术也难以造福患者。”在访谈过程中,李政道再三提起“安全”“有效”两个词,表达建立严格质控体系的必要性。也正是基于对质量安全的极致追求,李政道决定将自己50多年的研究和管理经验系统化,打造一套再生医学的“基础设施”。这个“基础设施”的核心,是一套“三位一体”的质量管理体系。何为“三位一体”?首先是AABB协会认证体系。该体系被誉为行业质量金标准,通过认证企业的产品质量可以获得全球认可。AABB认证的细胞临床实验室,将成为本项目质量管理体系中的框架和基石。其次,尤为值得一提的是李政道独创的质量管理体系,一种在AABB框架下的KNOW HOW管理方式。国内再生医学公司的质量管理文件只有操作流程部分(一般为5-30页)。而李政道带领团队在此基础上不断迭代更新,将操作流程细化增补到1000页以上,形成千页级操作规范,在确保更佳可执行性的同时,还从源头上确保合规。此外,本项目的质量管理体系中,有AI程序系统对再生医学整套流程进行数据管理,以进一步提升产品质量。03从实验室到临床的完整链条规划建设亚洲最严谨的再生医学临床实验室外,项目群的另一个目标便是形成从基础研究到临床应用的完整链条,即从基础研究开始,通过科研成果落地转化,延展至产业发展。其中,基础研究将重点发力干细胞和外泌体在免疫性疾病、老年病等领域的应用。在临床转化环节,在拥有建设AABB标准的实验室经验后,一个符合AABB标准的干细胞回输中心也有望面世。而在链接产业和自我造血方面,项目群还将开展CRO/CDMO业务,助力行业产品研发与认证。而这一多元的业务组合有望在开展过程中形成闭环。项目群当前针对老年群体面临的两大健康问题:一个是关节退化导致的行动不便,另一个是口腔功能的衰退,使老年群体难以享受美食。两者都严重影响了晚年生活质量。据悉,针对后者李政道及团队正在与北京大学等高校合作,开发针对老年人口腔功能修复的产品。而项目群引入的AABB标准,正是为了帮助中国再生医学与国际接轨。未来,随着更多再生医学药品获批入市,全球市场规模将快速扩张,这将推动国内监管政策跟进,形成相关市场发展的重要动力。多重力量将推动国内再生医学迎来行业级别的淘金热。04写在最后:步履不停公众了解李政道,更多是源自人教版课标本第七册《语文》中,一篇题为《跨越海峡的生命桥》的课文。1998年,也是在他的推动下,浙江大学医学院第一附属医院完成了第一例骨髓捐赠移植成功,开创了海峡两岸骨髓移植成功的先例,也成为更多人认识“血清干细胞之父”李政道的一个契机。事实上,在两岸未通航的年代,李政道已持礼遇签证往来百余次,以护送配型骨髓,挽救了上百人的生命。课文中记载的,对于李政道而言,或许只是尘世里数个日夜中的一个。如今,李政道再度启程,并直言看到了再生医学带来的改变,与团队推动再生医学项目群。无论是 AABB 协会的认证,还是李政道自身的质量管理咨询,CRO 或 CDMO 业务、再生医学大数据还是AI,都是淘金热中的卖铲子业务。不仅现金流好,利润好,而且容易形成行业级别的品牌影响力,增长性亦可期。现阶段,团队正在积极触达政府基金,并有条不紊地推进项目进一步落地。目前项目群正在寻求政府基金合作,如果您对项目群感兴趣,请与我们联系。*封面图片来源:123rf如果您想对接文章中提到的项目,或您的项目想被动脉网报道,或者发布融资新闻,请与我们联系;也可加入动脉网行业社群,结交更多志同道合的好友。近期推荐声明:动脉网所刊载内容之知识产权为动脉网及相关权利人专属所有或持有。未经许可,禁止进行转载、摘编、复制及建立镜像等任何使用。动脉网,未来医疗服务平台
免疫疗法细胞疗法
分析
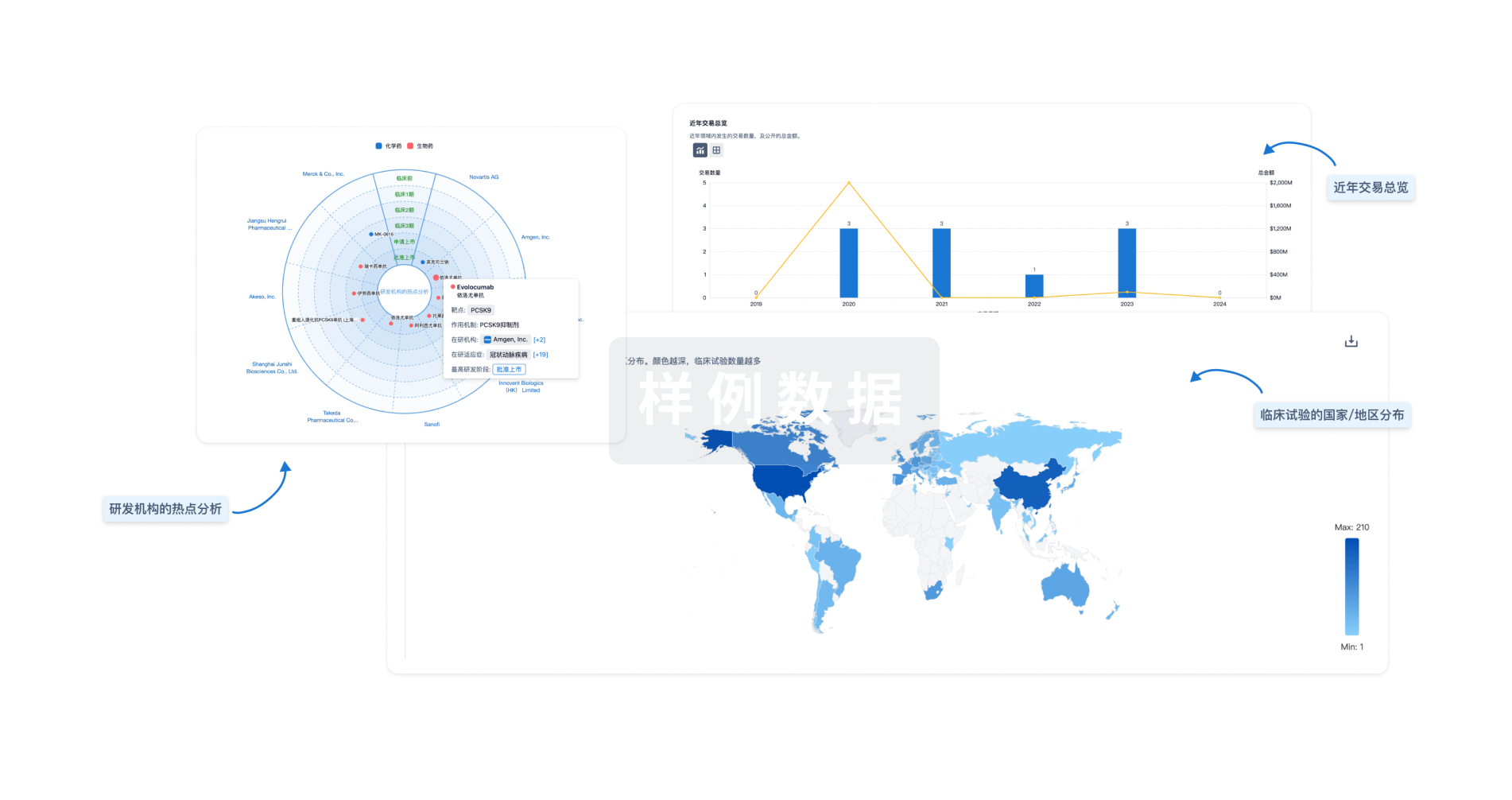
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用