预约演示
更新于:2025-06-25

Shanghai Bovo Biotechnology Co., Ltd.
更新于:2025-06-25
概览
疾病领域得分
一眼洞穿机构专注的疾病领域
暂无数据
技术平台
公司药物应用最多的技术
暂无数据
靶点
公司最常开发的靶点
暂无数据
| 疾病领域 | 数量 |
|---|---|
| 感染 | 7 |
| 排名前五的药物类型 | 数量 |
|---|---|
| 预防性疫苗 | 6 |
| 结合疫苗 | 2 |
| 重组载体疫苗 | 1 |
| 重组多肽 | 1 |
| 减毒活疫苗 | 1 |
| 排名前五的靶点 | 数量 |
|---|---|
| HPV L2(乳头瘤病毒小衣壳蛋白) | 1 |
| SARS-CoV-2 antigen(新冠病毒抗原) | 1 |
关联
7
项与 上海博沃生物科技有限公司 相关的药物靶点- |
作用机制 免疫刺激剂 |
在研机构 |
原研机构 |
在研适应症 |
非在研适应症 |
最高研发阶段临床1期 |
首次获批国家/地区- |
首次获批日期- |
靶点- |
作用机制 免疫刺激剂 |
在研机构 |
原研机构 |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
靶点- |
作用机制 免疫刺激剂 |
在研机构 |
原研机构 |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
4
项与 上海博沃生物科技有限公司 相关的临床试验NCT05672966
A Phase I, First-in-human, Randomized, Observer-blinded, Placebo-controlled, Dose Escalation Study to Evaluate the Safety, Tolerability, and Immunogenicity of BV601 in Healthy Adult Volunteers
This is a Phase I, first-in-human, randomized, observer-blinded, placebo-controlled, dose escalation study to evaluate the safety, tolerability, and immunogenicity of BV601 (a HPV Vaccine) in healthy adult volunteers.
开始日期2023-08-01 |
申办/合作机构  武汉博沃生物科技有限公司 武汉博沃生物科技有限公司 [+2] |
NCT05718037
A Phase I, Randomized, Observer-blinded Study to Evaluate the Safety, Tolerability, and Immunogenicity of BV211 in Adult Volunteers
This is a phase I, randomized, observer-blinded study to evaluate the safety, tolerability, and immunogenicity of BV211(a herpes zoster vaccine) in Adult Volunteers.
开始日期2023-08-01 |
申办/合作机构  武汉博沃生物科技有限公司 武汉博沃生物科技有限公司 [+1] |
NCT05706324
A Single-arm, Open-label Phase I Clinical Study to Evaluate the Safety, Tolerability and Immunogenicity of Recombinant COVID-19 Vaccine (Adenovirus Vector) for Inhalation in People 21 to 65 Years Old (Previously Primed With Authorized Vaccines)
In this trial, a single-arm, open-label study design will be used to evaluate the safety and tolerability after vaccination with escalating doses of the investigational vaccine (Recombinant COVID-19 Vaccine (chimpanzee adenovirus vector) for Inhalation, RCVi) at low, medium, and high doses in healthy adults (previously primed with authorized vaccines).
开始日期2023-06-01 |
申办/合作机构  武汉博沃生物科技有限公司 武汉博沃生物科技有限公司 [+2] |
100 项与 上海博沃生物科技有限公司 相关的临床结果
登录后查看更多信息
0 项与 上海博沃生物科技有限公司 相关的专利(医药)
登录后查看更多信息
11
项与 上海博沃生物科技有限公司 相关的文献(医药)2025-06-06·NUCLEIC ACIDS RESEARCH
Boosting CRISPR/Cas12a intrinsic RNA detection capability through pseudo hybrid DNA–RNA substrate design
Article
作者: Liu, Yi ; Qiao, Bin ; Jiang, Qingyuan ; Zhang, Junqi ; Qiao, Jie ; He, Ruyi ; Jin, Shuqi
Abstract:
The CRISPR/Cas12a [clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 12a] system is known for its intrinsic RNA-guided trans-cleavage activity; however, its RNA detection sensitivity is limited, with conventional methods typically achieving detection limits in the nanomolar range. Here, we report the development of a “pseudo hybrid DNA–RNA” (PHD) assay that significantly enhances the RNA detection capability of Cas12a. The PHD assay achieves a striking detection limit of 7.7 pM using single CRISPR RNA (crRNA) and 33.8 fM using pooled crRNAs. Importantly, this assay exhibits ultra-high specificity, capable of distinguishing mutated RNA target sequences at the protospacer adjacent motif (PAM)-distal region. It can also detect ultrashort RNA sequences as short as 6–8 nt and long RNAs with complex secondary structures. Additionally, the PHD assay enables PAM-free attomolar-level DNA detection. We further demonstrate the practical utility of the PHD assay by successfully detecting miR-155 biomarkers and human pappilloma virus 16 DNA in clinical samples. We anticipate that the design principles established in this study can be extended to other CRISPR/Cas enzymes, thereby accelerating the development of powerful nucleic acid testing tools for various applications.
2022-02-01·The CRISPR journal4区 · 生物学
Strategies for Enhancing the Homology-Directed Repair Efficiency of CRISPR-Cas Systems
4区 · 生物学
Review
作者: Liu, Yi ; Zhao, Xueke ; Liu, Hui ; Sun, Wenli ; Yin, Wenhao ; Qiao, Jie
The CRISPR-Cas nuclease has emerged as a powerful genome-editing tool in recent years. The CRISPR-Cas system induces double-strand breaks that can be repaired via the non-homologous end joining or homology-directed repair (HDR) pathway. Compared to non-homologous end joining, HDR can be used for the treatment of incurable monogenetic diseases. Therefore, remarkable efforts have been dedicated to enhancing the efficacy of HDR. In this review, we summarize the currently used strategies for enhancing the HDR efficiency of CRISPR-Cas systems based on three factors: (1) regulation of the key factors in the DNA repair pathways, (2) modulation of the components in the CRISPR machinery, and (3) alteration of the intracellular environment around double-strand breaks. Representative cases and potential solutions for further improving HDR efficiency are also discussed, facilitating the development of new CRISPR technologies to achieve highly precise genetic manipulation in the future.
2020-12-01·Biotechnology for biofuels1区 · 工程技术
Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethanol production
1区 · 工程技术
ArticleOA
作者: Ma, Lixin ; Sun, Wenli ; Liu, Yi ; Chen, Lixia ; Li, Shuntang ; Qiao, Jie ; Wang, Xinping ; Wu, Ke ; Dong, Ce
Abstract:
Backgrounds:
Engineering yeast as a consolidated bioprocessing (CBP) microorganism by surface assembly of cellulosomes has been aggressively utilized for cellulosic ethanol production. However, most of the previous studies focused onSaccharomyces cerevisiae, achieving efficient conversion of phosphoric acid-swollen cellulose (PASC) or microcrystalline cellulose (Avicel) but not carboxymethyl cellulose (CMC) to ethanol, with an average titer below 2 g/L.
Results:
Harnessing an ultra-high-affinity IM7/CL7 protein pair, here we describe a method to engineerPichia pastoriswith minicellulosomes by in vitro assembly of three recombinant cellulases including an endoglucanase (EG), an exoglucanase (CBH) and a β-glucosidase (BGL), as well as a carbohydrate-binding module (CBM) on the cell surface. For the first time, the engineered yeasts enable efficient and direct conversion of CMC to bioethanol, observing an impressive ethanol titer of 5.1 g/L.
Conclusions:
The research promotes the application ofP. pastorisas a CBP cell factory in cellulosic ethanol production and provides a promising platform for screening the cellulases from different species to construct surface-assembly celluosome.
47
项与 上海博沃生物科技有限公司 相关的新闻(医药)2025-03-13
·同写意
中国的新药公司当下最看重的是BD和融资,而决定BD和融资的关键就是临床数据。因此,一个首席医学官(CMO)的实力对公司的商业化能力十分重要。
同写意曾两次盘点中国新药公司的CMO。之所以如此,是因为中国新药越来越多的进入了临床阶段,而且创新程度越来越高,不断向FIC和BIC发展,这就使得临床试验的复杂性越来越高,因此作为一个新药公司的CMO,其专业性也就显得尤其重要。特别是,这是这个公司的CEO对临床试验驾驭能力不够的情况下更是如此。
从本次盘点的近200位CMO信息来看,CMO是一个流动性较大的岗位,多数是在Biotech公司之间流动,黑永疆、何如意博士则在Biotech和传统药企之间切换岗位。少数的CMO成长为公司的CMO或自己创业发展,比如邹建军和回爱民博士,我们列出了11位。
需要说明的是,本次盘点的近200位现任CMO的信息我们没有做一一核对,一定会有人已经离开了表中所列的岗位,就请大家留言板留言更正吧。
本文整理自网络信息,未经企业核实,实际内容请以官方为准。
189位
首席医学官
按姓氏首字母排序
01
Adrian M. Di Bisceglie
君圣泰CMO
2019年7月出任君圣泰CMO。曾任美国肝病研究协会的前任主席和理事会成员、AASLD杂志《肝病学》的编辑委员会成员,并担任圣路易斯大学胃肠道和肝病分部的内科教授和肝病科主任,并于2006-2017年担任圣路易斯大学内科系主任。
02
昂秋青
曙方医药CMO
昂秋青博士拥有超过18年的临床开发丰富经验,曾在赛神医药、科伦药业、礼来、辉瑞、GSK担任首席医学官、全球临床科学负责人、医学总监和临床研究医生等职位。
03
Bijoyesh Mookerjee
先声药业CMO(肿瘤领域)
加入先声之前在诺华担任肿瘤医学副总裁兼高级全球项目临床负责人。在此之前,曾在葛兰素史克、因塞特医疗、阿斯利康、托马斯杰斐逊大学西德尼·金梅尔癌症中心、马里兰大学格霖邦癌症中心和约翰斯霍普金斯肿瘤中心等国际领先医药企业与医疗科研机构担任管理及科学岗位。
04
蔡建明
兆科眼科CMO
2021年6月,蔡建明加入兆科眼科,出任公司CMO。曾任PPD亚太区副总裁及医学总监、MDCO的亚太区副总裁;也曾任职于多间全球领先的药业公司,包括Bayer Schering Pharma AG、默沙东、Amgen及GE Healthcare等担任高级医学职位。
05
蔡胜利
维立志博CMO
加入维立志博前,蔡博士曾任恒瑞(美国)副总裁,负责全球肿瘤项目的临床开发。之前,先后在德国拜耳全球肿瘤临床开发部、日本第一三共公司、瑞士诺华肿瘤和美国Intrexon Corporation担任临床研发项目负责人。
06
蔡东坡
尚德药缘CMO
2012年至2015年担任哈佛大学医学院华人专家学者联合会副主席,负责转化和精准医学活动。拥有多年中国临床经验并了解中国临床体系。拥有接近完整的跨越基础研究,转化医学和临床一期,二期的抗癌药物研发经历。
07
陈兆荣
多玛生物CMO
曾在祐和,琅铧医药,喜康生物医药,Sanofi,Bayer,GSK等多家国内外知名企业就职,做过医学研究,各期药物研发,药物审评,药物注册,和药物安全等工作。
08
陈长汀
合源生物CMO
2021年7月,陈长汀博士加入合源生物。2005-2018年,担任FDA生物制品评估和研究中心(CBER)组织和先进治疗办公室(OTAT)医学官/团队负责人。
09
陈杰
驯鹿生物CMO
从事肿瘤临床医生工作14年。加入驯鹿生物前,曾任北恒生物CMO、武田中国副总裁兼医学事务主管,此前,还曾在BMS中国担任高级医学事务总监、辉瑞中国担任医学事务总监、三维生物担任高级科学家。
10
陈霞
泰格医药CMO
现任泰格医药CMO、高级副总裁。之前曾先后担任北京协和医院临床药理学研究中心副教授,北京天坛医院临床药理学教授,国家药品评审中心(CDE)临床评审员等职务。
11
陈剑锋
宇耀生物CMO
曾服务罗氏、葛兰素史克、诺和诺德以及江苏豪森、和记黄埔、科望等国内知名药企,任职涵盖医学总监、临床运营负责人和临床开发负责人,在临床医学和运营团队的组建和管理、临床开发策略制定与实施等方面拥有丰富经验。
12
陈泓恺
安立玺荣创始人兼CMO
回国前,陈博士任职于美国加州大学旧金山分校格拉斯通研究所担任助理教授。之后曾服务于葛兰素史克等多家生物医药研发机构,带领从早期研发到临床试验多项神经疾病领域新药的项目研发。
13
陈炳官
新为医药CMO
1990-1996年在日本川崎医科大学附属病院、姬路坂本病院等从事外科临床;1996-2008年在美国密歇根大学医学院、耶鲁大学医学院;2008年回国至今分别在上海东方医院、合肥京东方医院等单位。同时兼任同济大学医学院外科教授,兼任东方医院中心实验室主任5年。
14
迟海东
礼来中国CMO
2010年,加入礼来公司任礼来中国副总裁兼CMO。曾就职于葛兰素史克中国及中国研发中心,历任医学顾问,临床研究医师,资深临床研究医师等职。
15
Caroline Germa
创胜集团全球药物开发执行副总裁及CMO
曾就职于阿斯利康、百时美施贵宝、诺华、辉瑞、法国礼来及赛诺菲-安万特。法国巴黎及里尔大学医学博士学位和医学肿瘤学家学位,以及乳腺疾病和免疫学硕士学位。
16
Daniel Canafax
凯瑞康宁CMO
2018年担任凯瑞康宁CMO,领导着从概念、临床前研究、临床研究到市场批准以及上市后研究的临床药物开发。此前,他曾在Omeros、Theravance、ARYx Therapeutics、XenoPort等担任要职。
17
Dr Richard Jones
和美药业CMO
曾在Roche、Johnson&Johnson、InventivHealth等多家国际医药公司及CRO公司担任关键岗位。原百明信康(WORG Pharmaceuticals)创始成员&CMO。
18
邓承军
华汇拓医药CMO
2020年12月起任上海华汇拓医药副总经理兼CMO。之前,曾担任福建金乐医药学术总裁(2012年9月-2017年7月)和北京夸克侠医学副总经理(2017年8月-2020年12月)。
19
David Margolis
腾盛博药CMO
加入腾盛博药前,Margolis博士曾担任GSK和ViiV Healthcare的医学总监和高级医学总监,并作为首席医师负责长效整合酶抑制剂cabotegravir临床开发计划。
20
杜云龙
普祺医药CEO/CMO
现任普祺医药CEO兼CMO、合伙创始人,从事药物研发、临床开发和产品上市全过程及管理逾25年,具有辉瑞、GSK等著名跨国制药公司20余年工作经历。
21
Erin Quirk
拓臻生物总裁兼CMO
2019年加入拓臻生物任CMO,Erin Quirk曾任吉利德临床研究副总裁、默克研究实验室临床研究总监。
22
Frederick Herman Hausheer
乐普生物Global CMO
2020年10月任乐普生物Global CMO。曾任曾担任药明康德全球CMO兼高级副总裁;及曾担任BioNumerik Pharmaceuticals创办人、董事长兼总经理23年。
23
Gerry Cox
北海康成首席战略官兼代理CMO
2019年4月GERALD COXD出任北海康成CSO和代理CMO。在创建自己的咨询公司前,曾担任过Editas Medicine的CMO,还曾在赛诺菲担任罕见病临床开发副总裁等岗位。
24
甘黎明
瑞博生物CMO、全球研发总裁、联席CEO
加入瑞博生物前曾任担任阿斯利康心血管、肾脏以及代谢类疾病的全球临床开发负责人,自2009至今兼任Sahlgrenska大学医院心血管疾病的转化医学研究和药物开发教授、首席医生。
25
郭晓宁
纽福斯CMO
郭博士有10余年在跨国制药公司及全球CRO研发临床开发及投资组合管理的经验。加入纽福斯前,曾担任赛生医药副总裁、研发负责人兼CMO,全面负责公司多条创新药管线开发和上市后产品的生命周期管理。
26
顾薇
映恩生物CMO
曾在多家跨国药企(勃林格殷格翰,阿斯利康,百时美施贵宝)和本土制药公司担任临床研究及临床开发负责人,管理经验全方位涵盖临床开发各个领域,包括注册,医学,临床运营,数据统计及药物警戒等。
27
葛蓝
元羿生物CMO
先后在强生、灵北、拜耳、索元生物、PPD任职,历任亚太区和中国区临床开发负责人。
28
Htay Htay Han
三叶草CMO(疫苗业务)
曾任武田疫苗业务早期临床开发项目负责人。在此之前,她在葛兰素史克疫苗公司(GSK Vaccines)工作了23年以上,担任临床研发项目负责人、高级总监等职。
29
何正治
辉诺医药CMO
2021年加入辉诺医药,任CMO和首席策略官。曾在罗氏制药作为感染和免疫领域研发专家和研发领导小组核心成员工作20余年,并作为顾问为默沙东及歌礼制药工作数年。
30
何如意
扬子江药业CMO
曾担任荣昌生物CMO,曾在FDA工作逾17年,担任多个战略领导职务。在中国,何博士曾任CDE首席科学家,国投创新医疗健康首席科学家,也是同写意新药英才俱乐部荣誉理事长。
31
贺李镜
真实生物CMO
加入真实生物之前,曾在多家全球性医药企业担任高管并领导临床团队,曾任葛兰素史克中国研发总经理和CMO、诺华中国副总裁和CMO、安进(亚洲)亚太区研发总经理、和记黄埔医药资深副总裁和CMO、奕拓医药执行董事和CMO。
32
侯兴华
汇宇制药CMO
曾就职于辉瑞、阿斯利康、复星等本土领先药企和跨国药企,担任首席医学官、高级副总裁等职务,曾于中国医科大学附属第一医院从事多年临床工作。拥有10余年临床研发相关工作。
33
华续赟
傲意信息CMO
上海中医药大学附属岳阳医院骨科副主任医师。现任中国康复医师协会脑功能监测与控制专业委员会全国委员、青年副主任委员。此外,他还是教育部中医智能康复工程研究中心康复机器人实验室主任。
34
胡敏
前沿生物CMO
曾就职于上海华山医院、默沙东制药、拜耳医药,曾任拜耳医药医学与临床负责人。
35
胡国强
普米斯生物CMO
曾在辉瑞、勃林格殷格翰工作,且曾担任信达生物临床研发高级副总裁、上海倍而达药业执行副总裁兼CMO等岗位。
36
胡云富
泛生子CMO
曾在FDA担任职多年,主导批准了包括首例基于NGS的LDT伴随诊断服务、首例基于NGS的伴随诊断试剂盒、基于NGS的肿瘤大panel检测服务、首例NSCLC液体活检伴随诊断试剂盒等。
37
黄英杰
香雪生命科学CMO
曾任复星制药海外CMO。此外还曾先后在诺华,施贵宝,葛兰素等跨国制药企业以及亚盛,创胜等以中国为基地的生物技术公司,出任过临床开发和医学事务部门的管理岗位。
38
黄薇
石药集团 Oncology CMO
资深肿瘤内科医生,专注于全身系统治疗和放疗。先后供职于MerckSerono1.5年,Roche 7年,BeiGene 3年,Simnovabio 1年。
39
黑永疆
石药集团 Global CMO
曾任智康弘义联席CEO、再鼎医药CMO和誉衡生物CEO。曾先后在齐鲁药业、美国Ambrx、美国安进、安进中国、诺华、罗氏等跨国公司任职要职。
40
韩德平
百吉生物CMO
美国罗切斯特大学肿瘤免疫学研究员;6年肿瘤临床医生经验;20年癌症疫苗研究及T细胞疗法研发和临床研究经验。
41
James Mc Leod
应世生物CMO
曾任美国Galleon制药公司CMO及高级副总裁。曾先后担任先灵葆雅早期临床研究与试验医学部门副总裁、美国默克公司实验医学部门副总裁。还曾就职于瑞士诺华制药。
42
Jeysen Yogaratnam
药物牧场CMO
2021年5月出任药物牧场CMO。在全球肝病、流感、艾滋病和肿瘤临床药物开发和医学方面拥有20年的商业及学术经验。
43
嵇靖
和誉医药CMO
2021年2月加入和誉前,曾任联拓生物医学和临床开发高级副总裁,阿斯利康中国研发副总裁。此前,先后在强生中国、GSK、赛诺菲-安万特和默沙东等跨国公司担任研发重要职位。
44
蒋皓媛
养生堂集团CMO
养生堂上海分公司总经理。曾任恒瑞医药临床医学部执行总监,海康德保瑞医学科学事物部及临床运营部高级副总裁等职。
45
姜雁林
真兴医药CMO
曾在青岛国大生物等多家药企担任高管,系原国家医药管理局生物技术专家委员会委员、山东省医药生物技术学会副理事长、山东医科院特聘教授。
46
贾懿
泽辉生物CMO
贾懿博士是药物临床开发专家,外科医生,毕业于上海医科大学、伦敦帝国理工医学院和北京协和医学院。曾就职于拜耳、辉凌,并任艾尔建中国临床研发负责人。
47
金建军
舶望制药CMO
曾在惠氏、勃林格殷格翰和礼来中国担任临床开发和医疗事务治疗领域负责人,以及诺华中国的临床开发负责人。曾任中国一家 SiRNA初创公司的CMO,以及一家医疗保健投资公司的运营合伙人,并在中国成功孵化和成立了一家肾科初创公司。
48
康晓燕
齐鲁制药临床研发中心常务副总经理、CMO
曾在东方肝胆外科医院消化内科工作。2007年2月至2009年8月,在美国康奈尔大学医学院从事博士后研究工作,先后任职于拜耳、勃林格、江苏恒瑞、上海君实等企业。
49
Kelly McKee
博沃生物CMO
曾历任昆泰传染病与疫苗卓越中心疫苗与公共卫生科副总裁,昆泰公共卫生与政府服务科副总裁兼首席医学官/科学官,昆泰公共卫生与政府服务科副总裁兼总经理。
50
孔庆贤
纽瑞特CMO
2025年1月任纽瑞特CMO。曾任先灵葆雅医学事务经理,罗氏临床项目研究团队负责人拜耳医学总监,新基公司高级医学总监,诺华制药疗乳腺癌的 ribociclib和alpelisib产品全球医学事务负责人,恒瑞临床肿瘤副总裁;齐鲁制药CMO兼集团副总裁,驯鹿医疗/亦诺微医药CMO。
51
Lars E. Birgerson
阿诺医药CMO兼美国CEO
现任阿诺医药CMO及阿诺医药美国CEO。曾任BMS医疗研发资深副总裁。此外,他也曾在多家国际顶尖药企担任高级管理职位,包括罗氏旗下基因泰克及罗氏制药副总裁。
52
李浩
康龙临床CMO
德泰迈医药创始人/董事长,曾任泰格益坦创始人和董事总经理,泰格医药首席医学官;曾担任全球领先的医药企业:昆泰,雅培制药,赛诺菲巴斯德,施贵宝,和Amylin等国际公司总部的经理到资深医学总监。
53
李敏
泰尔康CMO
美国杜兰大学医学院教授,上海瑞金医院肿瘤质子中心合作交流部部长/研究员。曾是日本临床实验公司CEO。
54
李玮
琅钰集团CMO
曾在罗氏、辉瑞等多家知名药企担任要职,在加入琅钰集团之前担任罗氏制药医学部副总裁。
55
李东辉
爱科百发CMO
曾就职于日本理化研究所,担任抗体平台项目负责人。也曾就职于日本杨森、阿斯利康和安进公司,担任医学总监、临床医学项目负责人、新药开发战略负责人等职位。回国后曾就职于和铂医药,担任医学免疫领域负责人。
56
李兴海
海创药业CMO
海创药业联合创始人/董事/首席科技官。曾任美国默克公司高级科学家、上海睿智化学研究有限公司资深总监、阿斯利康上海新药研发中心总监。
57
李锡明
亿一生物CEO兼CMO
曾在中美的跨国药厂包括礼来药厂、拜耳美国研究中心、法码西亚药厂、罗氏药厂、绿叶制药、石药集团等工作了25年多,任DMO、医学临床研究部门VP和其他高级职务。
58
路程
基因启明CMO
暂无介绍
59
李继刚
诺思格CMO
曾先后在Strong Memorial Hospital从事肿瘤科研,在强生中国(西安杨森)医学部负责精神科和肿瘤药物在中国的临床I-IV期研究。2010-2017年在西安杨森。
60
李彤
嘉和生物CMO
曾任轩竹医药高级副总裁兼临床开发主管、杨森中国研发中心高级总监及临床开发部主管,并曾在默克担任医疗事务经理。
61
李文玲
亘喜生物CMO
曾任EXUMA生物技术公司CMO,还先后在辉瑞、赛诺菲、基因泰克以及四环医药等公司出任过临床开发和医学事务部门的管理岗位。
62
李晓燕
维眸生物CMO
2021年1月出任维眸生物CMO。此前,曾任艾尔建/艾伯维全球临床执行医学总监(Executive Medical Director)和辉瑞眼科临床医学总监。
63
李长青
埃格林医药CMO
曾任FDA高级医学审评官。在多家跨国药企领导过新药研发的工作。加入埃格林医药前,他曾负责国际CRO巨头精鼎医药亚太区临床、法规、市场准入等咨询业务。
64
李卫华
复融生物CMO
在国内大型三甲教学医院从事20余年医疗、教学和科研工作,曾任消化科教授。曾先后在瀚森制药、齐鲁制药和Biotech公司担任医学团队负责人和CMO。
65
李依霖
征祥医药CMO
在新药临床开发领域具有近20年的工作经历,曾任职于国内著名医药上市企业,主导过多个药物的临床试验设计与管理。曾作为项目临床研究质量控制负责人,参与多项北京市科委大型临床研究课题的管理。
66
梁津津
天广实生物副总经理兼CMO
曾担任Icon全球副总裁及中国区总经理、康德弘翼副总裁兼总经理,以及大昆泰卓越数据中心前高级总监及总经理。
67
刘富鑫
熙源安健创始人、CMO
在疼痛/神经领域深耕多年,曾任普洛药业CMO,全面领导医学事务、临床运营、注册以及上市后医学支持等管理工作,对中美两国临床研究和注册的经验颇丰。
68
刘平
福贝生物CMO
2021年任福贝生物CMO。曾任石药集团、科伦制药临床研发负责人。此外,他还曾在Ausa Pharmed、UCB、礼来等药企担任要职,曾任奥萨医药的研发总裁。
69
刘清
极目生物CMO
曾担任爱尔康大中华区的临床开发及医学事务负责人,爱尔康亚太区医学总监,以及艾尔建中国区眼科医学事务负责人,艾尔建亚太区青光眼科学负责人。
70
刘春
东方略CMO
曾就职于北大未名、康辰医药,先后负责业务商务拓展、企业战略发展、决策委总负责人及研究院副院长职位。
71
刘世洲
英百瑞CMO
曾任担任冠科美博高级医学总监、强生血液肿瘤临床研发部门负责人、BI高级医学专家和项目负责人,以及浙江大学转化医学研究院和浙江大学医学院附属第一医院的双聘教授。
72
刘艳
米度生物CMO
曾任影研医疗科技CMO,曾担任法国Median Technologies首席医疗官,及欧洲癌症研究与治疗组织(EORTC)转化研究、放射治疗和影像部门的负责人。
73
刘锐
永泰生物CMO
曾任北京肿瘤医院癌症生物治疗及诊断中心副主任。
74
刘昌东
安龙生物CMO兼高级副总裁
曾就职于诺华制药下属的子公司Alcon,担任全球临床开发负责人;担任过欧康维视生物的CMO和首席科学家,武汉东宇生物总经理以及艾尔康生物联合创始人兼CMO;曾担任齐鲁制药、丽珠制药单抗生物和百奥泰生物的CMO及高级副总裁。
75
刘述森
领星生物CMO
曾任任BMS中国真实世界证据与流行病学负责人, 默沙东药物流行病学亚太区负责人,赛诺菲中国RWE负责人,神州医疗副总裁兼首席医学证据官,CDE真实世界研究的外部专家,并且拥有在国家CDC工作六年。
76
卢启应
宜明昂科CMO兼高级副总裁
曾先后进入Roche、GSK、AZ和Pfizer中国研发中心等跨国药企工作,2015-2019年在Pfizer中国研发中心工作,2017年起担任肿瘤产品组中国临床开发负责人&总监。
77
陆雨芳
创响生物CMO
曾在Regeneron, Celgene, GSK, 和Eisai等公司负责药物开发和医学事务。加入创响前,她是NASDAQ上市公司Celldex的临床开发副总裁,领导其免疫管线的开发。
78
陆志红
挚盟医药CMO
曾任标新CMO、绿谷制药全球开发总裁,并在葛兰素史克、优时比、索尔维、先声药业等企业工作过。
79
骆天红
金赛药业CMO(非肿瘤领域)
曾任礼来亚洲内分泌领域医学总监,赛诺菲中国医学事务部负责人,百时美施贵宝中国大陆及香港地区医学部负责人。
80
Marc E. Uknis
泰飞尔生物CMO
曾任Gemini Therapeutics公司CMO,也曾在ViroPharma, Shire, CSL Behring,Roche等医药公司担任要职参与和领导新药临床开发。
81
Melissa Palmer
甘莱制药CMO
曾任武田制药肝病临床研发负责人,还曾在夏尔制药和Kadmon任职。
82
Michael Silverman
科岭源CMO
曾供职于多家生物医药企业,包括Biopure Corporation、Telor Ophthalmic Pharmaceuticals、Sandoz Research Institute等。
83
Mark Lanasa
百济神州高级副总裁、实体肿瘤CMO
曾在阿斯利康担任副总裁、肿瘤学后期开发全球临床负责人职务。
84
Mark J. Gilbert
药明巨诺高级顾问、代理CMO
曾任Juno Therapeutics CMO。之前,Gilbert博士还在Bayer Schering Pharmaceuticals、Berlex Pharmaceuticals、Schering AG和Immunex等公司担任领导职务。
85
Matthias Straub
领晟医疗CMO
2020年06月出任领晟医疗CMO。先后任职Ciba Geigy、苏威制药和雅培等跨国企业。
86
Mehrdad Mobasher
百济神州血液学CMO
曾先后在基因泰克、Corvus Pharmaceuticals和再鼎医药任职。
87
孟杰天
万邦医药CMO
暂无介绍
88
母生梅
信立泰CMO
曾任辉瑞(中国)高级总监,负责罕见病和疫苗领域的研发。2023年加入信立泰,负责公司的临床开发和医学事务。
89
毛力
贝达药业研发总裁兼CMO
曾任赛生药业CMO、中国生物制药CMO,贝达药业资深副总裁兼CMO、丽珠生物公司CEO兼CMO;先后担任美国强生公司及其他大型药企的高级管理职务。
90
门宇欣
海昶生物CMO
于2003年加入FDA,历任CDER临床药理高级、特级审批官和药物审批小组负责人职位。曾任天津医科大学临床医学/天津第二附属医院住院医生。
91
孟海津
优锐医药CMO
曾服务于科研机构、跨国药企、初创及合资药企(加州大学洛杉矶分校、中国科学院、辉瑞、百时美施贵宝、誉衡生物及上药博康)。曾任BMS中国早期临床开发负责人、誉衡生物副总裁兼CMO、上药博康中国首席科学官。
92
孟玲
维健医药CMO
加入维健医药前,曾在拜耳德国柏林总部担任心血管治疗领域的全球医学事务副总监。
93
苗博龙
星眸生物CMO
曾先后在西安杨森制药&诺和诺德医学部、国内领先生物制药企业三生制药集团历任医学事务官、药品国际营销业务部&临床研究部门总监。
94
Mythili Koneru
传奇生物CMO
曾担任Marker Therapeutics首席医学官、礼来公司肿瘤免疫学副总裁,加入礼来前,是纪念斯隆-凯特琳癌症中心的肿瘤学研究员。
95
Nageatte Ibrahim
信达生物肿瘤学CMO
拥有在默沙东和GSK公司超过11年的肿瘤领域工业界药品开发管理和9年医疗及医学院校工作经验。
96
NgocDiep T.Le
嘉晨西海CMO
曾先后担任Amgen医学总监、葛兰素史克医学执行总监、诺华制药医学执行总监、阿斯利康肿瘤免疫创新药副总裁等。
97
聂秀清
新码生物CMO
曾任上海中医药大学附属普陀医院主治医师;先后在多家国际知名上市制药企业担任重要管理岗位。
98
倪翔
开拓药业CMO
超25年新药开发/药物警戒/临床质量管理经验,曾在阿斯利康等世界领先的国际制药巨头以及数个创新型生物科技公司任职,曾领导6项全球NDA及众多IND获批、拥有全球药品监管部门沟通经验。
99
Phillip Dennis
天境生物CMO
曾任赛诺菲肺癌战略副总裁兼新型、抗体偶联药物全球项目负责人,阿斯利康肺癌战略副总裁兼全球临床负责人。Dennis博士还在美国NIC担任了14年的转化研究员。
100
Paul Woodard
以明生物CMO
曾在Amgen、基因泰克、Bellicum等公司担任要职。
101
彭彬
轶诺药业CMO
曾任瑞士诺华临床药理部高级研究员、美国GSK肿瘤临床研究临床药理部高级医学总监、诺华制药全球肿瘤转化医学上海中心负责人及执行总监、上海岸迈生物CMO。
102
秦续科
宜联生物CMO
曾担任科望医药CMO、阿斯利康的高级医学总监,以及礼来公司的全球消化道肿瘤学科带头人。
103
秦志杰
禾元生物CMO
曾先后担任美国加利福尼亚大学圣克鲁斯分校研究员、美国 TOSK 制药临床研究科学家、AnaSpec 生物公司资深科学家和企业发展策划专家、美国 ReLIA Diagnostic 公司医学和市场总监、丹诺医药临床医学副总裁。
104
秦宏
惠泰生物CMO
曾先后在葛兰素史克、凯因科技及中美华东制药等从事医学事务和临床开发工作。
105
Ramon Mohanlal
万春医药CMO
万春医药EVP兼CMO.Ramon Mohanlal博士曾就任于GSK,Pfizer, Vertex, Interleukin Genetics, Syntium, Novartis,以及AstraZeneca等在内的大型药企和生物技术初创公司。
106
Raffaele Baffa
科济药业CMO
曾在Ziopharm Oncology担任CMO和研发执行副总裁,曾在 Medisix Therapeutics 担任CMO,以及在夏尔制药担任全球临床开发部副总裁和肿瘤学治疗区负责人,在施维雅制药收购夏尔肿瘤学部门后,Baffa博士担任施维雅制药的CMO。Baffa博士还曾在辉瑞和赛诺菲等药企任职。
107
Richard Lee
科越医药CMO
曾在Alexion Pharmaceuticals担任血液学负责人。曾在拜耳、罗氏、Genentech、Millennium/Takeda和百时美施贵宝等制药和生物技术公司担任临床和医疗事务领导职务。
108
Robert Andtbacka
高诚生物CMO
曾任Seven and Eight Biopharmaceuticals的首席医疗官,Huntsman癌症研究所黑色素瘤和皮肤肿瘤项目副主任和黑色素瘤临床研究项目主任。他还是Vestan Medical Imaging的首席医疗官。
109
任明
喜鹊医药CMO
曾任挚盟医药任CMO、北京临床研究中心副总裁兼CMO,在此之前,他是北京宣武医院临床研究的特邀教授和项目管理员。
110
任伟
香港养和集团CMO
曾在亚盛医药,国亦生命,卡恩细胞生物等生物公司任职CMO等职位。
111
Roger Sidhu
道明生物CMO
曾任Brooklyn(现为Eterna)ImmunoTherapeutics首席医疗官;Roivant Sciences执行副总裁兼首席医疗官,Cell Design Labs首席医疗官等。
112
Sujata Rao
英矽智能CMO
曾任礼来旗下ARMO Biosciences公司的全球临床开发副总裁、百时美施贵宝肿瘤免疫领域医学负责人和肿瘤学科学顾问、安进旗下Onyx Pharmaceuticals高级医疗总监。
113
Steffen Heeger
辐联医药CMO
曾在大型跨国药企和新兴制药公司担任高级肿瘤学临床开发职位超过15年。加入辐联前,曾在在Selvita, S.A.、Pega-One担任CMO。
114
Sharon Moore
康缔亚CMO
曾任一家CRO的首席医学和安全官。
115
石明
和黄医药执行副总裁、研发负责人及CMO
曾在诺华工作十多年,担任过诺华全球项目临床负责人。之后在创胜集团担任执行副总裁,全球研发负责人/CMO。
116
沈阳
欧康维视CMO
曾任参天制药(中国)有限公司医学信息部门经理,上海罗氏制药医学信息经理。
117
史军
启元生物CMO
曾担任缔脉生物首席开发官。曾在罗氏上海创新中心负责早期临床开发和临床药理学近8年。回中国前曾在美国的Berlex, Aventis, Forest, Daiichi-Sankyo 和Allergan共20年。
118
史青梅
基石药业CMO
曾任科文斯高级医学总监,精鼎医药新加坡及中国办事处医学总监。
119
宋钦辉
长乘医药CMO
2024年4月加入长乘医药任CMO。2022年10月任华赛伯曼CMO,宋博士曾在美国诺华制药从事新药研发工作近10年。
120
宋文杰
艾博生物CMO
曾任先声药业临床开发部高级副总裁、基石药业临床开发部副总裁、诺华肿瘤临床转化中心全球项目负责人、强生亚太研发中心项目负责人、阿斯利康临床医学总监。
121
滕燕
同宜医药CMO
曾任剑桥制药CMO并曾在BMS、诺华、辉瑞等多家公司担任要职。
122
童岗
神领锐医药CMO
曾任美国BMS医学总监和高级医学总监、美国科文斯公司执行医学总监、美国灵北医药公司老年痴呆症药物研发项目负责人、武田医药公司亚洲开发中心临床科学部门负责人。
123
汪海丹
勤浩医药CMO
曾在诺华、药明巨诺、诺思格等国内外知名药企和临床CRO公司从事临床开发工作。在加入勤浩以前,在石药集团担任临床开发副总裁。
124
汪裕
劲方医药CMO
曾先后在三维生物、礼来制药、葛兰素史克和赛诺菲亚太研发中心担任肿瘤领域医学负责人。
125
王东亮
求臻医学联合创始人、副总裁兼CMO
曾任哈尔滨医科大学附属肿瘤医院肿瘤内科的副主任医师。
126
王莞梅
泰诺麦博CMO
曾担任辉凌医药中国研发总经理、基石药业临床副总裁等职,开发项目涉及抗病毒、肿瘤、自身免疫性疾病等治疗领域。
127
王婷婷
来恩生物COO & CMO
曾在罗氏诊断公司亚太总部肿瘤医学事务部门带领亚太医学团队。在Tessa Therapeutics临床医学部门领导了FDA IND临床的设计,申报和获批。
128
王卫军
精诚医药CMO
精诚医药CMO、医学情报中心负责人,中国医药生物技术协会皮肤整形与修复分会秘书长。曾任梅奥中国转诊办公室首席医学官,英国NHS胸外科执业医生。
129
王雪鹂
凌科药业CMO
曾在Seres Therapeutics担任临床开发高级副总裁,之前,也曾在夏尔制药(Shire)、百特奥尔塔(Baxalta)和武田制药(Takeda),诺和诺德(Novo Nordisk)担任全球临床开发负责人。
130
王宜
加科思CMO兼高级副总裁
胰腺癌的肿瘤专家,曾就职于美国华盛顿大学,担任癌症中心消化道肿瘤科主任、治疗策略开发项目部(一期临床)主任。
131
王开
鑫康合生物CMO
曾在强生、拜耳、礼来等多家跨国药企和国内生物制药公司任职多年。
132
王莉
轩竹生物副总裁、CMO
曾经任职于基石药业、誉衡生物,在生物药及小分子药物领域有深刻的临床经验。拥有18年三甲医院肿瘤临床医生工作经验。
133
魏晓雄
汉都医药总裁、CMO
曾就职于Medpace公司、美国FDA临床药理学办公室、国立卫生研究院(NIH)的国家肿瘤研究院。
134
吴穷
翰森制药CMO
曾在临床肿瘤学领域从业二十余年。现任中国临床肿瘤学会理事、中国抗癌协会癌症康复与姑息治疗专业委员会副主委。
135
吴奕涵
先声药业高级副总裁兼CMO
曾任瑞石生物CMO,在GSK伦敦总部和GSK中国研发中心从事研发和管理工作多年。曾在英国伦敦及苏格兰公立医院担任肠胃外科医生。
136
吴济生
泽璟制药董事、副总经理、CMO
曾任上海康德保瑞、康德弘翼CMO,美国ProsoftClinical公司首席运营官及高级副总裁,方达医药高级副总裁、临床总经理,美国Graceway制药公司执行产品研发总监等职位。
137
吴诸丽
复星医药全球创新研发中心副总裁、CMO
曾先后在拜耳、杨森、罗氏等跨国医药公司担任临床研发的相关职务。
138
王瑶
康朴生物CMO
曾在AstraZeneca、Allergan、Kyowa Kirin、Celgene和Amgen等长期从事临床研发工作。
139
王树海
迈威生物CMO、高级副总裁
曾任复星医药研究院院长助理、临床研究中心部长,上海信谊药厂医学总监、医学注册部及项目管理部部长。
140
王琳娜
中国生物制药,正大创新研发中心CMO
暂无介绍
141
危士虎
昆拓医药CMO
具有十多年药物和医疗器械开发经验、药企和CRO医学事务工作经验。
142
宛卉
西比曼CMO
曾在罗氏和武田等制药公司担任过多个领导职务,包括血液学全球医学总监和肿瘤学执行医学总监等。在武田公司担任新兴市场肿瘤学医学事务负责人。
143
Willard Dere
安济盛CEO首席顾问兼CMO
曾任美国安进CMO、全球临床开发负责人,美国礼来内分泌、骨及普药研发副总裁。之前,曾担任犹他大学内科教授、犹他大学健康科学中心临床和转化科学中心个性化健康执行主任和联合首席研究员。
144
肖申
海森生物CMO
曾任思路迪医药/海森生物CMO。曾任FDA临床高级审评员,负责FDA心、肾、血管疾病新药的临床审评。加入FDA之前,曾为内科临床主治医生。
145
肖东
睿诺医疗CMO
曾任邵阳医院心内科医师,海正药业临床试验研究首席科学家。
146
谢志逸
英派药业高级副总裁兼CMO
曾任亚狮康制药CMO,诺华肿瘤公司的医学顾问,台北退伍军人总医院桃园分院的主治医师。
147
徐志新
原启生物CMO
曾先后担任诺诚健华CMO和圆因生物CMO,也曾先后担任罗氏美国、安万特药业集团、辉瑞等知名药企的临床药理/转化医学部门高级总监。
148
徐应永
启函生物CMO
曾任再鼎医药医学副总裁,也曾就职于百时美施贵宝和新基医药。
149
徐俊芳
华东医药CMO
曾在阿斯利康、强生、赛诺菲等跨国医药公司担任临床研发的相关职务。
150
项安波
石药集团事业部总裁兼非肿瘤CMO
曾任创响生物CMO兼高级运营副总裁、齐鲁制药任CMO、恒瑞医药资深医学总监,并在葛兰素史克(日本)从事新药研发十四年。
151
石燕
启德医药CMO
石燕博士在肿瘤综合治疗领域拥有17年的临床治疗实践经验。曾任职于中国人民解放军总医院、上海交通大学医学院附属瑞金医院肿瘤科。
152
延锦春
恒瑞医药副总经理兼CMO
曾任康诺亚生物肿瘤CMO。曾在美国强生和拜耳制药工作;也曾担任美国安博生物CMO,并在美国华盛顿大学担任过住院医师和专科医生。
153
杨春旭
爱思迈生物联合创始人兼CMO
资深国际医药安全专家,拥有三十余年的国际临床治疗和医药研发经验,涉及药物上市前和上市后的药物警戒及医学安全监控领域。
154
杨军
维昇药业CMO
曾任礼来中国糖尿病领域高级医学总监。
155
杨赞东
璧辰医药CMO
曾任索元生物CMO、信达生物全球高级副总裁兼美国总经理、Spectrum制药公司全球临床研发高级副总裁,此前,还曾在Inovio制药公司、杨森生物/强生制药公司、诺华和默克等公司任临床研发项目负责人。
156
姚毅
世领制药CMO
曾任美国联邦政府高级医疗官。曾任美国FDA高级资深临床评审官。也曾任士泽生物CMO。现任澳门特首长官和政府医药评审顾问,中国工程院免疫学组外籍顾问。
157
于晓天
诺辉健康CMO
领导并完成5项国内首创三类体外诊断试剂的临床试验、注册准入与产品上市工作,发表多篇SCI论文。
158
于顺江
上海医药CMO
曾在恒瑞医药担任医学总监、资深医学总监,齐鲁制药肿瘤新药CMO,曾在香港和记黄埔医药(上海)担任高级临床管理岗位。
159
于沛川
靖因药业CMO
曾在Centessa Pharmaceuticals担任临床开发高级副总裁;在Global Blood Therapeutics(GBT)公司任职临床科学负责人及副总裁。在此之前,他曾分别在美国Portola制药,Alexion制药和吉利德科学担任临床开发管理职务。
160
余健鸿
冠科美博CMO
曾在FibroGen、Anesiva等公司任职。
161
岳勇
来凯医药CMO
曾在强生、赛诺菲及GSK任职,领导过多个肿瘤药的全球多中心临床试验上市。
162
姜一星
翊博生物CMO
马大癌症中心资深临床教授,曾主导多个临床试验,肿瘤学教科书编委。
163
金小平
科伦博泰CMO
曾任康方生物高级副总裁和临床开发部负责人,也曾先后在国际知名制药企业美国第一三共、美国阿斯利康等从事上市前药物临床开发工作。
164
翟一帆
亚盛医药CMO
曾担任美国国家癌症研究所外科分部的博士后研究员,此后历任GSK、Bayer、Exelixis、HealthQuest等多家公司。
165
张云
亚虹医药CMO
曾在恒瑞医药、赛诺菲巴斯德、武田制药等国内外知名药企担任过临床研究、医学事务和注册事务的高级管理职位。
166
张虹
甫康药业CMO
曾在皮尔法伯担任临床开发总监&中国医学总监,也是昆泰大中华区首席医学官。更早前,还分别在Parexel和QuintileIMS从事临床研发。
167
张建慧
瑞科生物CMO
曾就职于领军医药公司赛诺菲-健赞、默沙东中国研发中心、萌蒂中国、博纳西亚等,担任临床医学负责人及CMO等职位。
168
张磊
翰思生物CMO
曾任索元生物CMO,新基制药全球执行医学总监和计划负责人,诺华制药肿瘤学全球业务部临床适应症主管,也曾在礼来制药总部担任要职。
169
张灵
益方生物CMO、副总经理
曾先后任职于GSK、强生、赛诺菲、先灵葆雅、默沙东和第一三共等国际大型医药公司临床高管。
170
张静
朗来科技CMO
曾任艾斯拓康CMO、艾伯维临床研发执行总监、艾尔建临床研发执行总监和美国强生临床研发总监。还曾在FDA工作10多年,担任临床团队负责人和临床审评员。
171
张向阳
诺诚建华CMO
曾任Hengrui Therapeutics公司CEO及董事会成员,曾任GSK临床开发高级总监。早年还曾在BMS、强生等跨国制药公司工作。
172
张晓静
德琪医药CMO
曾任诺华制药肿瘤领域部分产品的医学负责人,在拜耳医药全球研发中心历任资深医学专家、肿瘤领域部分产品全球临床研发负责人。也曾担任恒瑞医药子公司副总经理、恒瑞CMO(肿瘤)兼副总经理。
173
张文君
昕传生物联合创始人、CMO
曾担任国内知名三甲医院的血液临床研究中心副主任,长期从事血液肿瘤的免疫治疗以及白血病干细胞靶向干预的基础和临床研究。
174
张善中
星奥拓维执行总裁兼CMO
曾就职于GSK、赛诺菲健赞、住友制药。
175
郑盼
昂科免疫创始人、董事、CMO
美国马里兰大学医学院终身教授,获选美国医学与生物工程院(AIMBE)选为2022年院士。
176
赵燕
艾斯拓康CSO & CMO
曾担任诺华全球药品开发(中国)首席战略与创新官,诺华全球药品开发(中国)负责人,诺华肿瘤药品开发及医学事务副总裁等职位。在加入诺华前,先后服务于瑞士罗氏制药、BMS。
177
赵荣华
复诺健CMO
曾担任上海三维生物医学部主任,主导了溶瘤病毒创新药物H101和H103的临床前和临床研究。
178
赵家宏
盛世泰科CMO
复旦大学医学院生物化学与分子生物学博士,CDE外聘审评专家,江苏大学客座教授。曾任泰格医药医学部总监、济民可信与高博医疗首席医学官。
179
朱奇
安道药业CMO
曾在一家CRO企业任临床执行总监,曾在BMS总部BMS MI和Lantheus(BMS剥离的上市公司)任全球临床研发总负责人共12年,在美国百特公司任高级医学官。
180
朱伟
博锐生物CMO
曾任优时比(UCB)全球免疫临床研发团队负责人,此前,在赛尔基因(Celgene)、百时美施贵宝(BMS)、诺华(Novartis)等全球知名医药企业负责全球临床研发项目。
181
曾庆雯
云顶新耀CMO
曾担任BMS血液肿瘤治疗领域全球开发项目负责人,Celgene和BMS中国的临床开发负责人。
182
朱永红
岸迈生物CMO
曾任罗氏制药上海创新中心全球转化医学项目负责人、复宏汉霖医学负责人、日本武田制药芝加哥全球研发中心的高级医学总监、Genentech资深临床科学家、罗氏制药的Palo Alto研发中心的生物标志物和实验医学负责人。
183
郑劲草
锐格医药CMO
拥有超过20年的跨国药企和国内大型药企工作经验,先后就职于礼来亚洲公司、惠氏制药、阿斯利康、诺华制药、辉瑞(中国)研究开发有限公司、云恒医药和恒瑞医药。
184
张峣
腾盛华创CMO、腾盛博药医学开发副总裁
曾参与腾盛博药新冠中和抗体药物安巴韦单抗/罗米司韦单抗的研发和临床试验工作,并在多个公开场合分享了该药物的研发进展和临床数据。
185
赵婧华
复星凯特CMO
曾服务于西安杨森、诺华、默沙东等医药公司。担任临床研究监查员、项目经理、医学顾问、医学事务总监、市场总监商务事业部负责人,疾病领域全国医学负责人,企业CMO等多个职务。
186
张宪
世和基因CMO
曾在美国德克萨斯大学安德森癌症中心担任肿瘤内科医生,专注于肿瘤的临床治疗和研究。曾在多家知名基因检测公司担任高级管理职位,负责临床研究、产品开发和市场推广等工作。
187
张昕
辉大基因CMO
曾任美国PointLoma生物科学公司创始人兼CEO、武汉纽福斯生物董事及首席运营官兼CMO、上海复宏汉霖全球临床医学与事务副总裁等。也曾在默克、拜耳、百健等国际知名企业任职。
188
周明
博安生物CMO
曾担任国家药品审评中心(CDE)的审评员。
189
张秀军
循生免疫医学转化医学研究院院长、CMO
循生免疫医学转化研究院院长兼理事长,长期从事新药开发研究,,参与和主持多个国际创新药研究和临床开发并获得产品上市批件。
11位
从CMO到CEO/公司创始人
按姓氏首字母排序
01
华烨
烨辉医药创始人、CEO
2014年回国之后,任和记黄埔医药临床开发及法规注册部负责人、CMO、资深副总裁。
02
回爱民
惠正奇医药董事长兼CEO
曾任复星医药执行总裁、全球研发总裁兼CMO、复星全球合伙人,赛诺菲全球副总裁,上海市重点干细胞治疗实验室主任。
03
李新燕
芳拓生物联合创始人、CEO
曾任芳拓生物CMO后升任CEO。拥有超过20年的创新药物研发和管理经验,曾任科济生物资深副总裁、昆药集团CMO、泽生科技常务副总经理、CMO。
04
鲁薪安
希济生物创始人/CEO
艺妙神州/希济生物创始人,艺妙神州CMO。2015年,联合创办艺妙神州并担任公司研发和临床方向负责人。创立希济生物并担任CEO。
05
庞建宏
JP Medical Pty Ltd 创始人、CEO
曾任CMO后升任CEO。曾任澳洲Sirtex临床医学总监、上海绿谷、缔脉、亚虹、斯微及淂晖资深副总裁、CMO 、CEO 及远大医药海外CMO等职位。现任中国多家医科大学客座教授。
06
申华琼
纽欧申医药创始人、CEO
曾担任天境生物的首席执行官,中国强生杨森制药公司开发总负责,恒瑞医药的首席医疗官,还曾先后陆续在美国礼来公司、惠氏公司和辉瑞公司担任中枢神经系统药物开发的临床负责人。
07
史琳
灵迅医药董事长兼CMO
中欧生命科学联盟副理事长。曾任远大医药副总裁兼CMO,比利时Janssen R&D的法规事务部神经系统新药欧洲法务部负责人,布鲁塞尔大学医学院麻醉科principle scientist。
08
汪文
天宜康医药CEO
曾任驯鹿生物CMO并升任CEO,后又创立天宜康医药并担任CEO。曾在西比曼生物科技集团、药明巨诺、先声药业集团担任医学总监、临床开发高级总监、临床开发执行总监等职位。
09
杨建新
基石药业CEO兼研发总裁
曾任基石药业CMO后升任CEO。曾任百济神州临床开发部负责人及高级副总裁,美国科文斯肿瘤医学总监,辉瑞肿瘤生物标记物资深首席科学家,Tularik/Amgen公司研究科学家。
10
朱俊
复宏汉霖执行董事、CEO
2021年1月起担任复宏汉霖高级副总裁,同时担任CMO。2023年7月升任为公司CEO。加入复宏汉霖前曾任职佳生中国CEO,IQVIA全球副总裁,欧姆尼凯医药科技中国区总经理,艾昆纬项目经理。
11
邹建军
君实生物总经理兼CEO
曾任新基医药中国医学事务负责人、总监,恒瑞医药副总经理、CMO。在此之前,还曾担任拜耳医药临床试验医师、拜耳中国肿瘤治疗团队负责人、拜耳总部Xofigo的全球医学事务总监。
相关链接
2021中国新药公司首席医学官(CMO)名录
高管变更CSCO会议
2025-02-08
作者 | 吴克 博士(博沃生物创始人、CEO)
编辑 | 芝麻核桃
前言
2024年新年伊始,在瑞士达沃斯世界经济论坛上,一种被命名为“X”的未知病原体成为全球领导人和各行业领军人物热议的焦点。世卫组织介绍说,所谓“X”是“假设会出现”的一种引起全球大流行的病原体,它可能导致的死亡人数将达到新冠病毒的20倍,可能导致多达5000万人死亡。如今新冠已成为一种常见且反复出现的流行病。加强国际信息交流、开展合作防控是抗击“X”病原体的根本保证,而疫苗再度成为焦点,再次成为了人们期待可以有效应对未来疫情的手段之一[1]。
岁末年初,让我们先盘点一下疫苗行业在2024年有哪些重大事件。
一
价格调整
2024年5月,中国疫苗行业迎来了一场“价格地震”。四价流感病毒裂解疫苗的中标价格从128元/支降到88元/支,HPV疫苗价格也出现调整,沃森生物的双价HPV疫苗以单价27.5元中标。这一降价潮不仅涉及流感和HPV疫苗,还包括其他二类疫苗产品,平均降价超过30%。
图片来源:江苏省公共资源交易中心发布的《关于调整部分疫苗供应价格的通知》
这一变化背后,是疫苗行业在市场供需关系和政策导向下的自我调节。一方面,随着疫苗生产技术的进步和市场竞争的加剧,疫苗的生产成本逐渐降低,为价格调整提供了空间;另一方面,政府为了提高疫苗接种率,保障公共卫生安全,通过政策引导和采购机制,推动疫苗价格回归合理区间。这一“瘦身计划”虽然让部分企业面临短期的利润压力,但从长远来看,有助于扩大疫苗接种人群,提升疫苗行业的社会价值,也为疫苗行业的可持续发展奠定了基础。
二
WHO预认证
2024年8月,中国疫苗行业再添喜讯,沃森生物的双价人乳头瘤病毒疫苗和科兴生物的脊髓灰质炎灭活疫苗均通过了世界卫生组织预认证(WHO-PQ)。WHO-PQ是疫苗进入国际市场的重要“门票”,通过预认证的疫苗,意味着其在质量、安全性和有效性上达到了国际标准,能够在全球范围内被广泛认可和使用。这一成就的背后,是中国疫苗企业在研发、生产、质量控制等环节的不懈努力,以及中国疫苗行业监管水平的不断提升。
三
新冠疫苗自费了
2024年7月24日,黑龙江省公共资源交易网发布《黑龙江省新冠病毒疫苗准入评审结果公告》,披露了自费新冠疫苗的价格区间为126元~371.3/支[2]。客观地说,新冠病毒已成为呼吸道传染病的常见病原体之一,应当进行常态化防控,对于高风险人群而言仍然需要接种。据国内研究结果表明,约10%~30%的新冠感染者出现了长新冠(Long-COVID),症状的严重程度和发病风险都会随着再感染次数的增加而显著增加,而且感染新冠可能会促进对多种病原体的易感性。此外,感染新冠可能加剧了原有疾病或引发了慢性并发症[3]。因此,如何确保有接种意愿的人群能够接种到安全、有效的新冠疫苗,并能够继续唤起公众对新冠感染防控的意识,是一个值得探讨的问题。
四
百日咳疫苗接种程序优化
针对近年来全球和我国百日咳疫情回升,我国小月龄婴儿和学龄儿童发病风险有所升高的情况,国家疾控局等6部门决定自2025年1月1日起,在全国范围内将现行3月龄、4月龄、5月龄、18月龄各接种1剂次吸附无细胞百日咳-白喉-破伤风联合疫苗(以下简称百白破疫苗)和6周岁接种1剂次吸附白喉-破伤风联合疫苗的免疫程序,调整为2月龄、4月龄、6月龄、18月龄、6周岁各接种1剂次百白破疫苗的免疫程序。此次调整不仅是传染病防控大势所趋,也是国家对百日咳疾病防控的有力举措[4]。
接下来,我们将分别从研发、技术、市场、投资等各个角度进行年度回顾,分析疫苗行业面临的机遇和挑战,探讨行业未来的趋势。
PART.01
研发篇
2024年,我们目睹了新技术的曙光与重磅产品的崛起。
mRNA疫苗技术继续保持其“明星光环”,并不断拓展应用领域。自新冠疫情期间崭露头角后,mRNA疫苗凭借其独特的作用机制和高效的研发速度,成为疫苗行业的热门领域。Moderna和BioNTech等公司布局了一系列mRNA疫苗管线,涵盖呼吸道病毒、慢病毒和其他类型的病原体。
2024年7月,GSK以4.3亿美元的价格收购了CureVac公司的候选mRNA流感疫苗和新冠疫苗的全部权益。
2024年9月11日,GSK发布了全新的2期临床试验数据,显示优化后的mRNA疫苗在年轻人和老年人中均能产生与常规流感疫苗对甲型流感和乙型流感相当的免疫反应[5]。
8月17日,辉瑞和BioNTech公布其第二代三价流感mRNA疫苗Ⅱ期临床试验的最新进展。该试验显示了令人鼓舞的数据,与常规流感疫苗相比,该疫苗对所有病毒株均显示具有强大的免疫原性。
9月5日,《细胞》发布了Moderna抗猴痘病毒的mRNA候选疫苗mRNA-1769在预防猴子感染后发生严重疾病及降低体内病毒水平方面,表现显著优于目前已获批的猴痘疫苗[6]。
然而,Moderna旗下两款呼吸道合胞病毒(RSV)疫苗的临床试验在2024年12月被美国FDA叫停,令这一技术在该赛道的应用前景蒙上了一层阴影。FDA对Moderna的RSV mRNA疫苗的安全性提出质疑,称在候选疫苗mRNA-1345和mRNA-1365中观察到重症下呼吸道感染病例的不平衡,并暂停所有针对婴幼儿的RSV疫苗临床试验,这一消息导致Moderna当天股价大幅下挫9%[7]。
mRNA疫苗技术面临着疫苗的稳定性、递送效率和免疫原性等问题。多价mRNA-LNP疫苗存在mRNA编码抗原的异源多聚化问题,不同成分单体抗原组成的多聚体可能导致蛋白质错误折叠,从而无法如实展示正确的表位。多价疫苗中的一种成分还可能会对其他成分产生免疫优势,每种成分在低剂量时是否能诱导出有效的抗体仍需探索[8]。
纳米疫苗等新型疫苗技术在2024年也取得了突破。国家纳米科学中心研究员赵潇等人设计并构建了具有不同表面抗原图案参数的纳米新冠疫苗,在小鼠模型中发现这种纳米疫苗在体液免疫方面的效果与mRNA疫苗相当,并且具有更强的细胞免疫激活能力[9]。
图片来源:公开资料
多联多价疫苗的研发和应用成为2024年的亮点之一。例如,肺炎球菌结合疫苗的研发不断升级,辉瑞、默沙东、赛诺菲和GSK等企业均在开发更高价次的疫苗产品。9月4日,美国疫苗公司Vaxcyte宣布,其候选31价肺炎球菌结合疫苗VAX-31在50岁及以上健康成人的Ⅰ/Ⅱ期临床试验中取得了积极结果,在所有研究剂量中均显示出良好的耐受性,具备与20价疫苗相似的安全性,其独有的11种血清型则达到了优效。据该公司新闻稿,VAX-31可覆盖超过95%的美国50岁及以上成人中流行的血清型,有潜力在当前PCV疫苗的基础上增加12-40%的覆盖范围[6]。
在2024年,我们还惊奇地得知:一位自德国马格德堡的62岁男性被发现在2年半内接种了217剂mRNA新冠疫苗!经调查,该个体虽经过度接种,但并无不良反应,对新冠的免疫力提高,零感染,其对其它病原体的免疫力未受影响[10]。这件事提示我们,对于人体的免疫系统和机制,我们还有大量的未知领域有待探究。
PART.02
技术篇
2024年9月刊《生物技术与生物工程》杂志系统总结了疫苗工艺技术近十年的发展情况[11]。新冠大流行使得mRNA及病毒载体等新平台得以年产数十亿剂疫苗。分析手段、设备及生物工艺技术的进步,使得疫苗开发速度与生产规模都创造了新高。大分子结构功能表征技术,结合建模及数据分析,能以单个粒子的分辨率定量评估疫苗配方,并指导疫苗药物物质及药物产品设计,在精确评估疫苗关键质量属性方面发挥了重要作用。
无标记(label-free)及免疫测定技术,有助于抗原位点表征以及疫苗体外效力检测,通过与下一代测序等技术相结合,将加速疫苗的表征与上市。用于实时监控及优化工艺步骤的过程分析技术,使“质量源于设计”的原则得以实施,并加快了疫苗产品上市。
连续流生产、一次性工艺、无细胞技术、过程分析技术及放行检测方面的创新,有助于降低疫苗成本并提高效率。创新技术也正在取代传统批放行试验,以消除实验动物的使用。疫苗分析领域取得了巨大技术飞跃,影响了疫苗生产各个方面,提高了产品的一致性、生产效率、产品质量、产品检测速度及运营规模。
PART.03
市场篇
在国际市场,跨国疫苗生产商如辉瑞、默沙东等“强者恒强”,它们凭借其强大的研发实力、成熟的生产工艺和广泛的市场渠道,长期以来在全球疫苗市场中占据重要份额。辉瑞的Prevnar系列肺炎疫苗和默沙东的Gardasil系列HPV疫苗在全球范围内广泛使用,为公司带来了丰厚的利润。这些疫苗不仅在发达国家市场占据主导地位,还在新兴市场国家逐步扩大市场份额。跨国巨头凭借其品牌优势和产品质量,赢得了全球消费者的信任和认可。当然,随着新兴市场国家疫苗产业的崛起,跨国巨头也面临着来自新兴市场的竞争压力。
在人口出生率下降和消费疲软的背景下,疫苗行业面临市场竞争加剧的局面。随着全球疫苗市场的不断扩大和竞争的日益激烈,疫苗企业必须在技术创新、产品质量和市场拓展等方面不断提升自身竞争力,以应对来自同行和市场的双重挑战。
2024年,一些重磅疫苗产品延续放量趋势,如带状疱疹疫苗和肺炎疫苗等二类苗产品批签发同比快速增长。国药中生集团、华兰疫苗、北京科兴等头部企业凭借技术和市场优势占据较大市场份额。近年来,随着国内疫苗市场的不断扩大和政策支持力度的加大,本土疫苗企业迎来了快速发展的机遇,通过不断加大投入,在稳固国内市场的同时,开始向国际市场拓展,“出海”成为了众多中国疫苗企业的共同选择。
国内企业出海模式趋于多元化,包括成品出口、技术转移、本地化生产和合作研发等多种方式,这样不仅有助于企业拓展国际市场,还能更好地适应不同国家的市场需求和政策要求。作为东南亚人口最多的国家,印尼对疫苗的需求巨大,因而成为中国疫苗企业出海的重点区域。
与跨国巨头相比,本土企业在国际市场上的份额仍然较小,需要在技术创新、品牌建设和国际市场拓展等方面继续努力。
尽管目前全球疫苗市场在整体药品行业中所占比例较小,但其增长速度显著。新兴市场国家如中国、印度、巴西等国家拥有庞大的人口基数和不断增长的中产阶级,对高质量疫苗的需求日益旺盛。随着经济的发展和医疗水平的提升,对疫苗的需求呈现出快速增长的趋势。例如,中国在新冠疫苗研发和生产中展现出强大的能力,不仅满足了国内的需求,还向全球多个国家提供了疫苗支持。随着越来越多的中国疫苗通过WHO预认证,中国疫苗产品的出口有望加速。印度则在疫苗生产领域具有成本优势,印度血清研究所是全球最大的疫苗生产商之一。
2022年中国疫苗市场(不含新冠疫苗)规模达到850.7亿元,2023年中国疫苗市场(不含新冠疫苗)规模增长至1017.7亿元。中投产业研究院预计,2024年我国疫苗市场规模可达到1,245亿元,年均复合增长率约为21.21%,2028年将达到2,687亿元[12]。
PART.04
投资篇
从历史上看,对传染病疫苗公司的投资只占新兴生物制药资金总额的一小部分。在过去10年中(包括新冠大流行期间的投资高峰期),为生物制药公司筹集的风险投资总额中,只有3.4%流向了拥有传染病疫苗项目的公司。相比之下,肿瘤药物开发公司获得了38%的风险投资,比疫苗公司多12倍。尽管需要更多投资来应对未来和新出现的大流行威胁,以及区域性流行病,但通过疫苗科学的创新已经取得了很大成就。过去十年疫苗领域的创新在I期批准的临床成功率高于平均水平,疫苗为11%,而新型小分子仅为5.6%[13]。
根据联合国儿童基金会的报告估计,疫苗产品开发平均需要11.9年时间,投资约8亿美元才能将一种产品推向市场,而只有22%的候选产品能够成功完成临床试验。但是,疫苗在免疫方面的投资回报价值很高,仅以儿童生存率和经济回报来评估时,其价值往往难以衡量。基于“疾病成本”方法,每投资1美元用于疫苗接种,可获得26.35美元的投资回报。采用“统计生命价值”方法,得出的投资回报率更高:每投入1美元用于免疫,可获得约52美元的投资回报[14]。
弗诺斯特沙利文咨询合伙人毛化在2024年某次行业会议上的演讲指出:疫苗是一个“小而美”的行业。在疫苗行业里,市场逻辑主要看接种,发达国家接种率比较高之后,接种潮流会移到中等收入的国家,再到低收入国家,永远是一个增量的市场。另外还有技术的创新迭代。相对而言,疫苗市场的增长逻辑相对更加持续。中国疫苗人均消费水平较低(7.1美元),仅相当于美国(59.5美元)的1/8,日本(28.1美元)的1/4和欧洲五国(法德意西英17.4美元)的一半,说明国内接种率还有提升的巨大潜力。
中投产业研究院发布的《2024-2028年中国疫苗市场投资分析及前景预测报告》指出,受到新型疫苗研发和新兴市场如中国、印度及俄罗斯需求增长的推动,全球疫苗市场预计将在2030年达到1292亿美元,期间年复合增长率可达10.8%,显示出强劲的增长势头[12]。
然而,回望2024年,从场内基金的表现来看,全年跌幅最大的板块就是生物疫苗类基金,下跌幅度全部超过了17%,跌幅最大的是招商基金公司管理的疫苗龙头ETF,下跌了27.8%。疫苗股的下跌除了投入疫苗研发导致成本增加、新疫苗开发无力以及销售回款情况不佳、计提坏账的原因外,可能也与新生儿的出生数量减少以及人们在疫情期间对疫苗的印象有关[15]。中国资本市场这样的表现,无疑为疫苗行业的发展带来了隐忧。
PART.05
挑战与机遇
01
研发成本高:技术创新的“拦路虎”
疫苗研发需要投入大量的人力、物力和财力,成本高昂。从基础研究到临床试验,再到最终获批上市,一款疫苗的研发周期通常长达数年甚至十几年。新冠疫苗的研发虽然在短时间内取得了突破,但背后是全球科学家和企业的共同努力,以及巨额的资金投入。疫苗作为具有极强公共属性的产品,我们期待着政府和国资能够更多地关注和支持这个战略新兴行业,也期待着真正的“耐心资本”能够伴随企业成长。
02
分销和接种难度大:最后一公里的“瓶颈”
在一些地区,由于基础设施不完善、卫生资源有限等原因,疫苗的分销和接种面临较大的困难。特别是在偏远地区和低收入国家,疫苗的冷链运输和储存条件难以满足要求,导致疫苗接种率较低。例如,非洲部分国家的疫苗接种率远低于全球平均水平,主要原因之一就是疫苗配送和接种环节存在问题。如何提高疫苗的覆盖率和接种率,是疫苗行业需要解决的重要问题之一。此外,欧美的研究都表明,电子信息的推送可以有效提高疫苗的接种率[16、17]。这也提示全社会特别是政府机构应当加大宣传推广疫苗的力度。
03
新技术的应用:人工智能助力疫苗研发
新技术的应用为疫苗研发提供了新的思路和方法。例如,利用人工智能和大数据技术可以通过分析大量的生物数据,预测病毒的变异趋势,提高研发效率,优化疫苗的抗原设计,从而提高疫苗的保护效果。
04
全球合作与政策支持:携手应对公共卫生挑战
根据国际货币基金组织报告,新冠疫情导致2020年全球GDP下降6.5%,预测到2025年将造成全球总计28万亿美元的经济损失。世界卫生组织2024年5月21日发布的最新版《世界卫生统计》报告显示,在2020年和2021年,新冠疫情分别导致了410万人和880万人死亡,新冠大流行在短短两年内使全球近十年来在提高预期寿命方面取得的进展化为乌有,退回到了2012年的水平[18]。
面对全球性的公共卫生挑战,各国政府和企业纷纷加强合作,共同推进疫苗研发和生产。例如,在新冠疫情期间,全球多个国家和企业通过“新冠疫苗实施计划”(COVAX)合作,加速疫苗的研发和分配。同时,各国政府也加大了对疫苗行业的政策支持力度,通过补贴、税收优惠等方式,鼓励企业加大研发投入,提升疫苗生产能力。这种全球合作和政策支持的模式,为疫苗行业的发展提供了良好的环境。
05
公共卫生投资增加:筑牢健康防线
预计全球将增加对公共卫生基础设施的投资,包括疫苗接种计划,以提高对未来疫情的准备和应对能力。公共卫生投资的增加,不仅有助于提升疫苗接种率,还能改善全球卫生系统的整体水平。例如,一些国家正在加大对疫苗冷链运输和储存设施的投资,以确保疫苗在运输和储存过程中的质量不受影响。这种投资的增加,将为疫苗行业的发展提供更广阔的空间。
PART.06
趋势篇
随着医疗技术的不断发展,个性化疫苗和精准医疗将成为疫苗行业的重要发展方向。个性化疫苗是根据个体的基因特征和免疫状况,定制适合个体的疫苗产品。精准医疗则是在疾病诊断和治疗过程中,根据患者的个体特征,制定精准的治疗方案。个性化疫苗能够提高疫苗的免疫效果,降低疫苗接种后的不良反应,为个体提供更精准的保护。2024年,个性化疫苗在肿瘤治疗领域取得了初步进展。研究人员通过对患者肿瘤细胞的基因测序,分析其突变特征,然后设计出针对这些突变的个性化疫苗。这种疫苗能够特异性地激活患者的免疫系统,攻击肿瘤细胞,从而实现对肿瘤的精准治疗。虽然目前个性化疫苗还处于临床试验阶段,但其展现出的巨大潜力已经引起了广泛关注。与此同时,个性化疫苗的研发和生产成本较高,需要在技术研发、生产工艺和质量控制等方面进行创新和突破。
随着基因测序技术、基因编辑技术、生物信息学等领域的不断发展和成本降低,个性化疫苗和精准医疗有望在未来几年内实现商业化应用,为疫苗行业的发展注入新的活力。
我们预测全球疫苗市场未来可能出现的趋势还包括:
技术进步和创新
随着新技术平台的发展,预计将有更多的疫苗快速开发并推向市场,特别是在面对新发和重新出现的疾病时。
供应链的区域化
受到新冠大流行期间全球供应链中断的影响,各地区可能会推动本地和区域生产能力的增强,以减少对外部供应商的依赖。
价格分级和可负担性
预计将继续存在不同收入群体之间的疫苗价格差异,但可能会有更多努力来减少这种差异,并提高疫苗的可负担性,特别是在中等收入国家。
成人疫苗市场的增长
随着人口老龄化和对成人疾病预防意识的提高,成人疫苗市场可能会迎来快速增长。大流行经验强调了全球合作的重要性,预计未来将有更多的国际协议和合作机制来应对全球公共卫生紧急情况。
全球合作和公共卫生安全
大流行经验强调了全球合作的重要性,预计未来将有更多的国际协议和合作机制来应对全球公共卫生紧急情况。
数字健康和疫苗追踪
数字技术的应用,如区块链和数字追踪系统,会在疫苗市场中发挥更大作用,以确保疫苗的可追溯性和减少假冒伪劣疫苗的风险。
疫苗接种的公平性
全球疫苗接种的公平性将成为重要议题,特别是在提高低收入和中等收入国家疫苗获取能力方面。
环境和气候变化的影响
气候变化可能会影响疫苗需求的模式,例如通过改变疾病传播的地理范围,这可能会促使市场对某些疫苗的需求增加。
政策和监管环境的变化
随着全球对疫苗重要性认识的提高,可能会出现新的政策和监管措施,以支持疫苗的开发、批准和分发。
公共卫生投资的增加
预计全球将增加对公共卫生基础设施的投资,包括疫苗接种计划,以提高对未来疫情的准备和应对能力。
结 语
在变革中前行
疫苗是解决传染病威胁的有力手段,是人类最伟大的公共卫生成就之一。据美国疾病控制和预防中心(CDC)估计,在1994-2023年出生的儿童进行常规疫苗接种,将预防约5.08亿例疾病、3200万例住院和1,129,000例死亡,直接节约了5400亿美元的开支,全社会开支的节约高达2.7万亿美元[19]。《柳叶刀》2024年5月发表的研究显示,疫苗接种在过去几十年中避免了1.54亿人死亡,其中5岁以下儿童1.46亿名,疫苗接种对降低全球婴儿死亡率的贡献占到了40%。这一数据充分证明了疫苗在公共卫生领域的重要价值[20]。
世卫组织《2030年免疫议程》明确提出,疫苗接种要覆盖从婴幼儿到老年的整个生命周期,在生命任何阶段不让任何人掉队。儿童疫苗的优先级一直高于专为成人设计的疫苗。儿童免疫计划包括明确的指导方针、明确的免疫计划和入学要求,这些因素共同推动了学龄儿童疫苗接种率达到90%。相比之下,成人疫苗往往面临认知度有限、可及性、可负担性和疫苗犹豫等挑战。
2020-2021和2021-2022年度我国流感疫苗全人群接种率分别为3.16%和 2.47%;在免费政策支持下,老年人接种率分别为42.71%和32.94%,明显低于WHO提出的重点人群流感疫苗接种率达到75%的目标。我国老年人肺炎球菌疫苗接种1.23%~42.10%,接种率水平低于美国(2020年为67.5%)。2018-2020年我国9~45岁女性人乳头瘤病毒(HPV)疫苗累计估算接种率为2.24% ,明显低于2018年美国19~26岁成年女性HPV疫苗的52.5%~53.3%接种率(至少接种1剂)[21]。
预计未来十年内,全球获批疫苗产品的数量将增长3倍,预计将有100至120种产品,用于预防40种不同的疾病。当前,成人疫苗主要分为两大类别:针对流感、肺炎、带状疱疹和新冠等广为人知的疾病的疫苗,以及针对旅行或地方性疾病的疫苗,如乙型肝炎、埃博拉、黄热病、蜱传脑炎和日本脑炎。未来,成人疫苗将扩展至院内感染疫苗,如艰难梭菌、金黄色葡萄球菌等以及针对未被满足需求的疾病的疫苗。预计到2032年,美国每年的成人疫苗接种量将超过5亿剂,是目前水平的2.5倍[22]。
2024年12月19日,WHO官网发布了一年一度的疫苗市场报告《Global vaccine market report 2024》。2023年,几个重要疫苗在整个生命周期内的接种覆盖率均低于《免疫规划2030议程》(IA2030)设定的90%目标。麻疹疫苗接种覆盖率不足,导致过去五年来103个国家暴发疫情[23]。2023年全球麻疹病例激增至1030万例,比2022年增加20%。这提示我们:疫苗接种,任重而道远。
2024年,疫苗行业在内卷和变革的旋律中砥砺前行,从价格调整到管理模式的创新,从免疫接种受众的拓展到接种方式的革新,以及国际格局中中国疫苗地位的崛起与挑战,这一年既有重大突破的喜悦,也有挑战带来的焦虑。
展望未来,疫苗行业将继续在技术创新、市场拓展和国际合作等方面不断努力,为人类健康事业做出更大的贡献。
请记住——“疫苗不仅是预防疾病的工具,更是人类对健康的承诺。”在这个瞬息万变的时代,我们需要的是信心和定力。
参考资料
1. https://finance.sina.com.cn/jjxw/2024-01-19/doc-inaczekn8834566.shtml?r=0&tr=381
2. https://news.qq.com/rain/a/20250101A00C4K00
3. https://www-thelancet-com.libproxy1.nus.edu.sg/journals/lanwpc/article/PIIS2666-6065(24)00217-7/fulltext
4. https://news.qq.com/rain/a/20241227A077XR00
5. https://www.thepaper.cn/newsDetail_forward_28728849
6. https://finance.sina.com.cn/jjxw/2024-09-19/doc-incprwmw3905292.shtml
7. https://finance.eastmoney.com/a/202412173270904432.html
8. https://www-nature-com.libproxy1.nus.edu.sg/articles/s41573-024-01042-y
9. https://news.sciencenet.cn/htmlnews/2024/11/533999.shtm
10. https://www-thelancet-com.libproxy1.nus.edu.sg/journals/laninf/article/PIIS1473-3099(24)00134-8/fulltext
11. Buckland B., et al. Biotechnology and Bioengineering. 2024; 121(9Sep): 2543-2980 https://analyticalsciencejournals-onlinelibrary-wiley-com.libproxy1.nus.edu.sg/doi/epdf/10.1002/bit.28703
12. https://baijiahao.baidu.com/s?id=1806148473112243401&wfr=spider&for=pc
13. https://www.bio.org/sites/default/files/2024-01/The-State-of-Innovation-in-Vaccines-and-Prophylactic-Antibodies-for-Infectious-Diseases.pdf
14. https://www.unicef.org/supply/media/17871/file/UNICEF-Vaccine-Markets-Prioritizing-and-Scaling-Up-Towards-Equitable-Access.pdf
15. https://baijiahao.baidu.com/s?id=1819967762370956913&wfr=spider&for=pc
16. Nature. 2024 Jul; 631(8019):179-188.
17. JAMA. 2024; 332(22):1900-1911.
18. https://www-who-int.libproxy1.nus.edu.sg/publications/i/item/9789240094703
19. https://www-cdc-gov.libproxy1.nus.edu.sg/mmwr/volumes/73/wr/mm7331a2.htm?s_cid=mm7331a2_e&ACSTrackingID=USCDC_921-DM133631&ACSTrackingLabel=This%20Week%20in%20MMWR%3A%20Vol.%2073%2C%20August%208%2C%202024&deliveryName=USCDC_921-DM133631
20. Lancet. 2024 May 25;403(10441):2307-2316.
21. 中国健康促进与教育协会.成人预防接种服务专家共识(2023年版)[J]. 中华预防医学杂志, 2024, 58(3): 275-284. DOI:10.3760/cma.j.cn112150-20240129-00098
22. Jones CH, et al. Exploring the future adult vaccine landscape-crowded schedules and new dynamics. NPJ Vaccines. 2024; 9(1): 27. doi: 10.1038/s41541-024-00809-z
23. https://cdn-who-int.libproxy1.nus.edu.sg/media/docs/default-source/immunization/mi4a/who_global_vaccine_market_report_2024_vdraft.pdf?sfvrsn=d7bffd94_5&download=true
作者简介
吴克 博士
博沃生物创始人、CEO
简介:上海市产业领军人才、湖北省政府特聘产业教授,中国疫苗行业协会理事、糖疫苗专委会副主委、国际合作促进委员会委员,中国食药促进会疫苗及生物制品分会副主委。
牵头主持国家重点研发计划、863项目、重大新药创制专项在内的多个项目开发,带领企业获评德勤“2020中国明日之星”、2023中国最具成长性疫苗企业TOP 10、2023亚洲及太平洋地区最佳疫苗品种AVEA大奖等。
声明:本文仅代表作者个人观点,不代表任何组织及本公众号立场,如有不当之处,敬请指正。如需转载,请注明作者及来源:蒲公英Biopharma。
推荐阅读
1、2024年中国生物行业主要政策回顾与盘点
作者:滴水司南
2、回顾展望生物医药:2024深度转型,2025破冰发展
作者:张金巍
3、2024年生物医药产业政策盘点(附政策包下载)
作者:芝麻核桃
4、生物医药产教融合:推动产业可持续发展的关键路径
作者:李峰
5、从"卡脖子"到"自主可控":生物医药供应链国产替代进行时
作者:杨清
6、生物制品抗体行业的2024
作者:黄庆
7、7大亮点回顾2024全球ADC产业图景
作者:药明合联
8、细胞治疗2024:盘点行业的破局之道
作者:张长风 博士
9、开启生命科学新纪元:2024基因治疗全景复盘
作者:马景梅
蒲公英生物运营团队恭贺新春
灵蛇献瑞 巳巳如意
疫苗信使RNA
2024-12-06
中文摘要
重组病毒载体疫苗是以病毒为载体递送目的抗原的一种疫苗。病毒载体技术路线早期多用于动物疫苗和难以用传统技术路线开发的病原体疫苗。我国针对新型冠状病毒感染疫苗研发布局的5条技术路线中,因重组病毒载体疫苗具备可同时产生较强的体液和细胞免疫、可通过黏膜途径接种以及1剂完成免疫程序等优势而获批上市,并在疫情防控中起到了重要作用。但重组病毒载体疫苗因预存免疫问题,即存在可能降低疫苗保护效力、为增强免疫原性可能导致安全性风险提升等因素,故上市的重组病毒载体疫苗产品较少。随着基因工程技术、分子生物学技术的发展,病毒载体种类日渐增多,并在多种疫苗中得到应用。此文就重组病毒载体疫苗的研发现状进行综述。
正文
重组病毒载体疫苗是将编码有效抗原的特定基因插入病毒载体并表达而获得的疫苗,可诱导体液及细胞免疫,病毒基因组稳定,无需佐剂即可诱导高免疫原性和持久免疫应答等,但存在预存免疫问题,当前研究主要通过人类稀有或灵长类动物腺病毒(adenovirus,Ad)来替代高流行性病毒载体、增加接种剂量等,来一定程度上克服预存免疫、加强免疫原性。病毒载体根据复制能力可分为2种,复制型病毒载体和复制缺陷型病毒载体。病毒载体疫苗平台是快速有效应对新突发传染病的途径之一,目前主要使用的病毒载体有Ad、痘苗病毒(vaccinia virus,VV)、疱疹病毒、水疱性口炎病毒(vesicular stomatitis virus,VSV)、流感病毒等。本文就各种用于预防或治疗不同疾病的病毒载体疫苗研究新进展进行综述。
1
病毒载体
1.1
双链DNA病毒
1.1.1 Ad载体 Ad直径70~90 nm,基因组约36 kb,在自然界广泛分布,人类Ad已有52种。Ad基因组包含早期表达的与病毒复制相关的E1—E4基因,第1代Ad载体剔除了E1和E3基因,缺失复制能力;第2代在第1代基础上删除E4基因,突变E2基因,降低了在293细胞中培养时产生复制型Ad的能力,虽提高了安全性,但降低了免疫原性;第3代为去除了所有编码蛋白质序列的空壳病毒,极大增加了载体容量,增加了安全性,也进一步降低了免疫原性,未规避预存免疫,病毒包装过程还需依靠辅助病毒和互补细胞系。重组Ad载体是非常有前景的载体之一,目前研究较多的有人5型Ad(Ad5)、26型Ad(Ad26)以及黑猩猩Ad(chimpanzee Ad,ChAd) 3型(ChAd3)等,用于已上市COVID-19疫苗、埃博拉病毒(Ebola virus,EBOV)疫苗等,以及临床研究阶段的呼吸道合胞病毒(respiratory syncytial virus,RSV)疫苗等。
1.1.2 VV载体 VV是一类体积大、结构复杂的病毒。病毒颗粒约为360 nm×270 nm×250 nm,基因组140~220 kb。VV宿主范围较广,作为载体可容纳大片段外源基因而不影响其生长特性。在开发靶向蛋白较大的疫苗时具有优势。如天花疫苗使用改良型VV安卡拉毒株(modified VV ankara,MVA),与天花病毒存在交叉抗原,能够表达有效抗原并诱导机体产生体液和细胞免疫,且安全性较好。VV载体被用于研究多种传染病预防,如EBOV疫苗、猴痘疫苗、流感疫苗等。
1.1.3 疱疹病毒载体 疱疹病毒的种类很多,共发现有100余种,人疱疹病毒直径120~150 nm,基因组120~150 kb,被开发为病毒载体的有单纯疱疹病毒(herpes simplex virus,HSV)、火鸡疱疹病毒、猪伪狂犬疱疹病毒等。疱疹病毒作为载体,可携带多个或较大基因,容纳外源基因能力强。其中,HSV能够感染多种细胞并快速增殖,利用这一特性改造的HSV基因工程载体可以用于基因和神经系统疾病治疗。2015年,FDA批准了第1个基于HSV-1的溶瘤病毒(oncolytic virus,OV)药物,该药物通过基因工程技术删除HSV-1基因组中的感染细胞蛋白(infected cells protein,ICP)34.5和ICP47基因,并将粒细胞-巨噬细胞集落刺激因子(granulocyte-macrophage colony-stimulating factor ,GM-CSF)插入ICP34.5位点,用于治疗黑色素瘤。HSV作为OV载体的研究热点,可发挥原位肿瘤疫苗功能,激活抗肿瘤免疫,多应用于肿瘤治疗。HSV-1和HSV-2作为载体均已被开发成OV候选药物,并且已进入临床试验阶段。
1.2
单链RNA病毒
1.2.1 仙台病毒(Sendai virus,SeV)载体 SeV又称乙型副流感病毒Ⅰ型,直径150~200 nm,基因组长约15 kb,是典型的人畜共患病毒。基于SeV载体的OV药物因潜在的癌症治疗作用而受到关注。SeV载体具有宿主范围广、表达量高、胞质表达、安全性好等优点,作为利用反向遗传学构建的新兴RNA病毒载体,SeV载体目前广泛用于基因治疗药物和新型疫苗研究。日本DNAVEC公司开发的用于治疗下肢缺血的SeV-hFGF2/dF,是国际上首个获批临床研究的SeV载体基因治疗药物。此外,应用SeV载体开发的HIV、HSV疫苗等处于临床前研究阶段。
1.2.2 麻疹病毒(measles virus,MV)载体 MV直径100~250 nm,基因组长约15 kb,可导致儿童急性呼吸道疾病。目前我国麻疹发病有低龄化趋势,且易伴有并发症。作为病毒载体,MV具有可容纳6 kb的外源序列并稳定表达、同时诱导产生显著的细胞和体液免疫应答、仅在胞浆中复制不会整合到宿主基因组等优点。通过将与目标病毒感染、复制或逃逸有关的蛋白基因插入MV反基因组载体中开发的HIV、寨卡病毒和基孔肯雅病毒的候选疫苗均已步入临床试验阶段。
1.2.3 VSV载体 VSV长度150~180 nm,直径50~70 nm,基因组约11 kb,通过昆虫叮咬和家畜传染,人感染后仅出现流感症状,但家畜感染后表现为口、蹄等水疱样病灶,肉、奶产量下降,造成畜牧业经济损失。VSV基因组能容纳多个外源基因;病毒复制多代次后性状保持稳定;不会整合到宿主细胞基因组;增殖滴度高,免疫原性好,不受宿主抗病毒载体免疫影响,可诱导较强的细胞免疫和体液免疫。基于VSV载体开发的EBOV疫苗已获FDA批准上市,保护率达到97.5%。
1.2.4 流感病毒载体 流感病毒通常呈球形颗粒,直径约100 nm,但新分离的毒株大多为丝状颗粒,长度能达到20 μm。人流感病毒分为甲、乙、丙型。通过反向遗传学技术,在流感病毒中插入外源基因构建流感病毒载体疫苗。流感病毒载体在人体内可引起体液、细胞及黏膜免疫,复制周期无DNA,不会整合到宿主细胞,还可通过更换亚型解决预存免疫问题。目前以流感病毒为载体开发的疫苗包括已进入Ⅲ期临床试验的COVID-19疫苗,以及早期研究阶段的HIV疫苗、RSV疫苗等。
1.2.5 新城疫病毒(Newcastle disease virus,NDV)载体 NDV颗粒呈多形性,直径120~300 nm,长约15 kb。不同NDV株的毒力不同,多数NDV载体疫苗选择La Sota缓发型弱毒株。构建NDV载体疫苗首先利用反向操作技术获得感染性病毒克隆,随后进行分子遗传操作。重组NDV活病毒载体遗传性状比较稳定,毒株间不易发生重组;病毒复制在胞浆内完成,不产生DNA,不会整合到宿主细胞基因组。NDV弱毒株疫苗可同时诱导全身体液免疫、细胞免疫和局部黏膜免疫,有较好的保护作用,已在临床试验中验证了其安全性,NDV已被应用于COVID-19疫苗的临床前开发等。
2
重组病毒载体疫苗
2.1
COVID-19疫苗
COVID-19可引起急性呼吸道传染病,目前已经上市的重组病毒载体COVID-19疫苗主要采用人或黑猩猩Ad载体,流感病毒载体等开发的COVID-19疫苗已进入临床试验阶段。
国内上市的COVID-19载体疫苗,采用人Ad5去除了E1和E3基因并导入新型冠状病毒(severe acute respiratory syndrome coronavirus 2,SARS-CoV-2)刺突蛋白基因,在感染细胞后可表达SARS-CoV-2 刺突蛋白,诱导体液和细胞免疫。但人Ad5在人群中流行率较高,可通过提高病毒滴度或者加大接种剂量来解决预存免疫。临床研究发现接种1剂COVID-19载体疫苗14 d后,重症保护率和总体保护率分别为96.0%、63.7%,安全性良好。英国牛津大学与阿斯利康公司合作开发了以ChAd为载体的疫苗AZD1222,在载体中导入COVID-19全长刺突蛋白,采用加大接种剂量的方式解决预存免疫问题。受试者接种2剂AZD1222,第2剂加强接种后至少14 d,疫苗总体保护率为70.4%。俄罗斯Gamaleya公司开发的Sputnik V COVID-19疫苗是一种基于重组Ad26和Ad5的组合载体疫苗,2个载体均可表达SARS-CoV-2刺突蛋白,分别使用2种重组载体进行初次、加强免疫,第1剂接种后21 d保护率达91.6%。由于人呼吸道中的抗体水平远低于外周血中,SARS-CoV-2在鼻腔感染和复制时就容易出现免疫逃逸,因此Chen等将改造双重减毒甲型流感病毒载体插入SARS-CoV-2受体结合域基因,开发了可经鼻腔喷雾接种的活病毒载体疫苗dNS1-RBD,该疫苗株可在上呼吸道有限复制,NS1基因缺失增强了流感病毒载体的安全性,且可诱导强效的抗病毒应答,在Ⅲ期临床试验中呈现出广谱保护率和安全性。
2.2
EBOV疫苗
EBOV是一种能够引起烈性传染病的病毒,有较强的传染性和最高达90%的病死率。基于重组病毒载体技术路线开发的EBOV疫苗众多,主要采用Ad、VV和VSV载体。
中国人民解放军军事科学研究院与天津康希诺生物股份公司开发的表达EBOV Zaire型(ZEBOV)囊膜糖蛋白(glycoprotein,GP)的Ad5-EBOV疫苗,可产生有效的细胞和体液免疫应答,于2017年在国内获批上市,增加Ad5-EBOV疫苗免疫剂量可在一定程度上克服载体预存免疫影响。Ewer等以灵长类动物ChAd3为载体来降低预存免疫,将Ad的GP基因替换成ZEBOV或EBOV Sudan型的GP基因,开发的复制缺陷型ChAd3- ZEBOV疫苗可诱导细胞免疫。在重组痘病毒载体的EBOV疫苗研发中,为提高有效抗原的表达量,减少预存免疫影响,免疫策略均为序贯免疫,即不同技术路线的疫苗按照一定时间间隔、剂次开展的接种,也称为异源疫苗接种。西安杨森制药有限公司采用2种EBOV疫苗Zabdeno(Ad26.ZEBOV)和Mvabea(MVA-BN-Filo)异源接种,临床研究表明,双剂量方案具有良好安全性,该方案于2020年获欧盟委员会批准上市。提高异源接种间隔时间,抗体有效浓度最高可保持1年。同源疫苗接种是指相同技术路线的疫苗按照一定时间间隔、剂次开展的接种,异源和同源Ad26、MVA载体EBOV疫苗方案都具有良好的耐受性,异源第2剂加强接种免疫后效果更好。美国默沙东公司Ervebo疫苗是在减毒VSV中插入EBOV表面抗原GP,保留了一定的复制能力,通过该病毒的轻度感染即可产生特异性抗体,该品于2019年获FDA批准上市。Grais等发现,Ervebo维持预防效果可达2年之久。以VSV为载体开发的重组VSV-ZEBOV疫苗在Ⅲ期临床试验中显示良好安全性,提高接种剂量可降低预存免疫、增加免疫原性。
2.3
HSV疫苗
HSV可引起人类多种疾病,如口唇部疱疹、腰腹部疱疹以及生殖系统疱疹等。在新生儿、严重营养不良和免疫功能低下者等人群中可引发危及生命的感染。从20世纪30年代开始,虽然尝试了灭活疫苗、蛋白质亚单位疫苗、减毒活疫苗和DNA疫苗等各种技术路线,但目前还没有获批上市的疫苗。
目前虽已发现HSV基因组的ICP34.5等基因表达的蛋白质会导致HSV的免疫逃逸并减弱细胞对HSV的清除,造成HSV基因组在感染细胞内持续存在,形成“潜伏”状态,但HSV影响潜伏的蛋白及其机制尚未被阐明。尽管已有将HSV改造成载体荷载免疫调节基因如GM-CSF的OV用于肿瘤的治疗,但用于健康人群的疾病预防仍具有潜在风险。因此,HSV疫苗的开发主要采用其保护性抗原,构建病毒载体、重组蛋白或者mRNA核酸疫苗等技术路线。
HSV蛋白GP〔如糖蛋白D(glycoprotein D,gD)〕、衣壳结构蛋白和披膜蛋白(如VP16)、非结构蛋白ICP27等能够诱导细胞免疫,可作为疫苗靶标的蛋白。由于HSV疫苗开发可能需要多蛋白协同发挥免疫效应,因此,多个外源基因的病毒载体SeV、Ad等成为目前开发HSV疫苗的选项。
以HSV阴性人群预防和阳性人群治疗为目标,Ren等采用复制缺陷型SeV,通过反向遗传学方法构建了2个重组病毒载体,1个编码HSV-2 gD(HSV-2-gD),另1个编码HSV-2 ICP27,在动物实验中显示出良好的体液和细胞免疫应答。通过2种携带HSV-2抗原的复制缺陷型病毒载体疫苗加强免疫,可较好地克服预存免疫,产生较强的特异性免疫原性并能够有效地预防HSV-2引起的生殖器复发感染。通过Ad构建编码HSV-2-gD的重组载体疫苗,在动物实验中显示出良好的体液免疫应答,也可一定程度上抑制HSV-2感染复发时的症状。
2.4
RSV疫苗
RSV是导致呼吸系统疾病的主要病原体,可引发各年龄段人群的下呼吸道感染,更是导致婴幼儿病毒性呼吸道感染住院的首要因素。免疫力低下或患有慢性基础病的老年人感染RSV的风险高并极易导致原有基础病加重。目前开发RSV疫苗使用的病毒载体有Ad、流感病毒等。
Salisch等使用人群中低流行性的Ad26和Ad35载体开发的RSV疫苗,动物实验显示能够诱导有效的免疫应答。婴儿免疫系统未发育完全,老年人免疫系统弱,均需要免疫原性更强的疫苗。武汉博沃生物科技有限公司开发的RSV疫苗采用Ad5载体,以融合蛋白作为RSV疫苗的主要靶标,可避免其他技术路线RSV疫苗缺点,如减毒活疫苗的毒力返祖、灭活疫苗可能诱导的抗体依赖的疾病增强效应,该产品已进入中试样品制备阶段。Lee等将RSV 的融合蛋白262—276蛋白表位插入到流感病毒载体构建的疫苗,动物实验显示可诱导产生保护性中和抗体。
2.5
HPV疫苗
HPV感染症状表现为寻常疣、尖锐湿疣等,分低危、高危型。临床已证实高危型HPV感染是引起宫颈癌和癌前病变最重要因素。用于研发HPV重组载体疫苗的病毒载体有Ad、MV等。
以Berna商业疫苗株MV载体开发的预防性HPV疫苗,动物实验显示可诱导特异性抗体。Gupta等基于MV开发的HPV疫苗已进入Ⅱ期临床试验。浙江普康生物技术股份有限公司开发的HPV16重组Ad载体治疗性疫苗已进展至Ⅰ期临床试验。法国Transgene公司开发的TG4001是基于非复制型、高度减毒的重组牛痘病毒MVATG8042载体、含有HPV16抗原(E6、E7基因)的重组病毒载体疫苗,目前已进入Ⅱ期临床试验,其联合程序性死亡配体1抑制剂Avelumab治疗晚期HPV阳性肿瘤患者,试验结果表明二者联合使用安全,且在HPV 16阳性肿瘤患者中产生抗肿瘤活性。中国生物技术股份有限公司研发的疫苗以HSV-1 OV为载体,在构建载体具有溶瘤活性的同时,表达HPV E6、E7等多个蛋白的多个抗原表位,可诱导产生持久的细胞免疫应答。
2.6
流感疫苗
流感病毒是引起呼吸道疾病的主要病原体,临床一般表现为轻症,即急性起病、发热、头痛等,重症病例可出现病毒性肺炎、继发细菌性肺炎甚至休克。目前开发的重组病毒载体流感疫苗主要采用Ad和VV载体。流感病毒根据血凝素(hemagglutinin,HA)和神经氨酸酶(neuraminidase,NA)的抗原性分为不同的亚型,通过调整HA和NA的组合可解决预存免疫问题。
HA是流感病毒表面重要的糖蛋白,唐昕莹选择人群血清阳性率较低的灵长类动物来源的Ad载体,以HA为靶基因,插入多个流感病毒抗原表位制备的流感疫苗,通过加强免疫降低预存免疫,该品在动物实验中展现出有效的保护效果。口服疫苗可作用于黏膜免疫系统,美国Vaxart公司的口服Ad5载体H1N1疫苗VXA-A1.1在Ⅱ期临床试验中显示出有效保护率和安全性。美国Altimmune公司的Ad5载体鼻腔接种流感疫苗NasoVAX,在免疫原性最强的剂量下,可诱导体液、细胞和黏膜的广泛免疫应答,具有良好的免疫原性及安全性。韩頔等以Ad5为载体,构建了表达SARS-CoV-2 刺突蛋白和H3N2 HA的重组双价非复制型Ad5载体疫苗HAdV5-S1-2A-HA,该品在小鼠体内诱导对SARS-CoV-2和H3N2流感病毒感染的特异性体液与细胞免疫,为预防COVID-19和流感的双价疫苗研发提供了基础。将流感病毒A/Wisconsin/67/2005株的核蛋白和基质蛋白构建到MVA上获得的重组病毒载体疫苗MVA-NP+M1已进入Ⅱ期临床试验,该品可诱导良好的T细胞免疫应答。
3
展望
重组病毒载体疫苗是新型基因工程疫苗研究的热点,虽具有可诱导多种免疫、表达稳定等优点,但已上市产品仍较少,主要是预存免疫问题,包括单一病毒载体疫苗应用于多次免疫接种程序或人群中高流行性病毒(如Ad)产生的预存免疫;为降低预存免疫,采用多样化病毒载体疫苗序贯接种时,诱发非特异性免疫;载体中插入大容量外源基因时,可能存在构建效率低、脱靶、耗时长等问题。未来开发病毒载体疫苗要突破的关键技术包括:联合基因编辑和同源重组等新技术,构建多样化、可容纳大容量外源基因的DNA病毒载体;利用反向遗传学技术等,拯救重组病毒构建多样化RNA病毒载体;形成较为全面的安全、高效表达的病毒载体库;采用免疫组学等新型免疫评价技术,发现并鉴定更有效的免疫原及作用靶点,进一步降低预存免疫问题。建立病毒载体疫苗研发平台,可快速响应新突发传染病,开发应对多领域各类疾病的治疗性疫苗,对保障人民健康、促进生命科学技术发展具有十分重要的意义。
作者
邢思熙1,2 许方婧伟1,2 张云涛1,2
1中国生物技术股份有限公司科研管理部,北京 100024;2新突发传染病新型疫苗研发全国重点实验室,北京 100024
通信作者:张云涛,
Email:zhangyuntao@sinopharm.com
引用本文:邢思熙, 许方婧伟, 张云涛. 重组病毒载体疫苗的研究新进展[J]. 国际生物制品学杂志, 2024, 47(5): 318-324.
DOI: 10.3760/cma.j.cn311962-20231205-00021
识别微信二维码,添加生物制品圈小编,符合条件者即可加入
生物制品微信群!
请注明:姓名+研究方向!
版
权
声
明
本公众号所有转载文章系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系(cbplib@163.com),我们将立即进行删除处理。所有文章仅代表作者观点,不代表本站立场。
疫苗
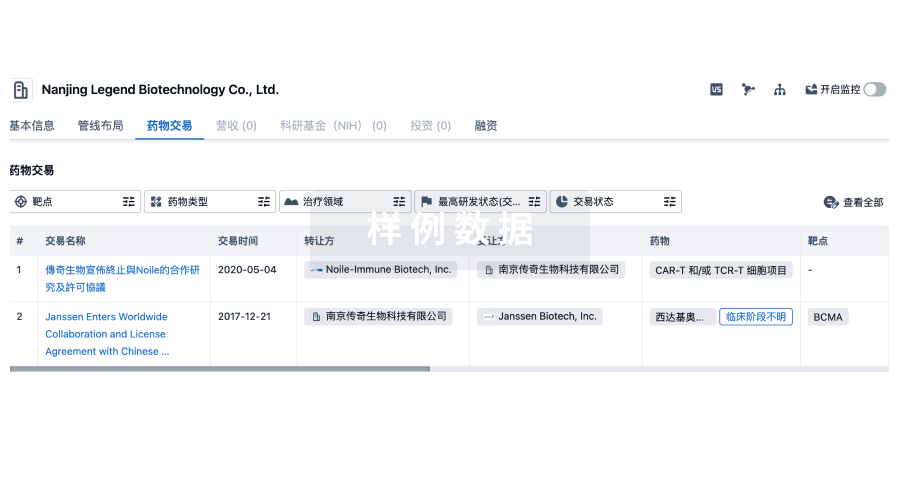
100 项与 上海博沃生物科技有限公司 相关的药物交易
登录后查看更多信息
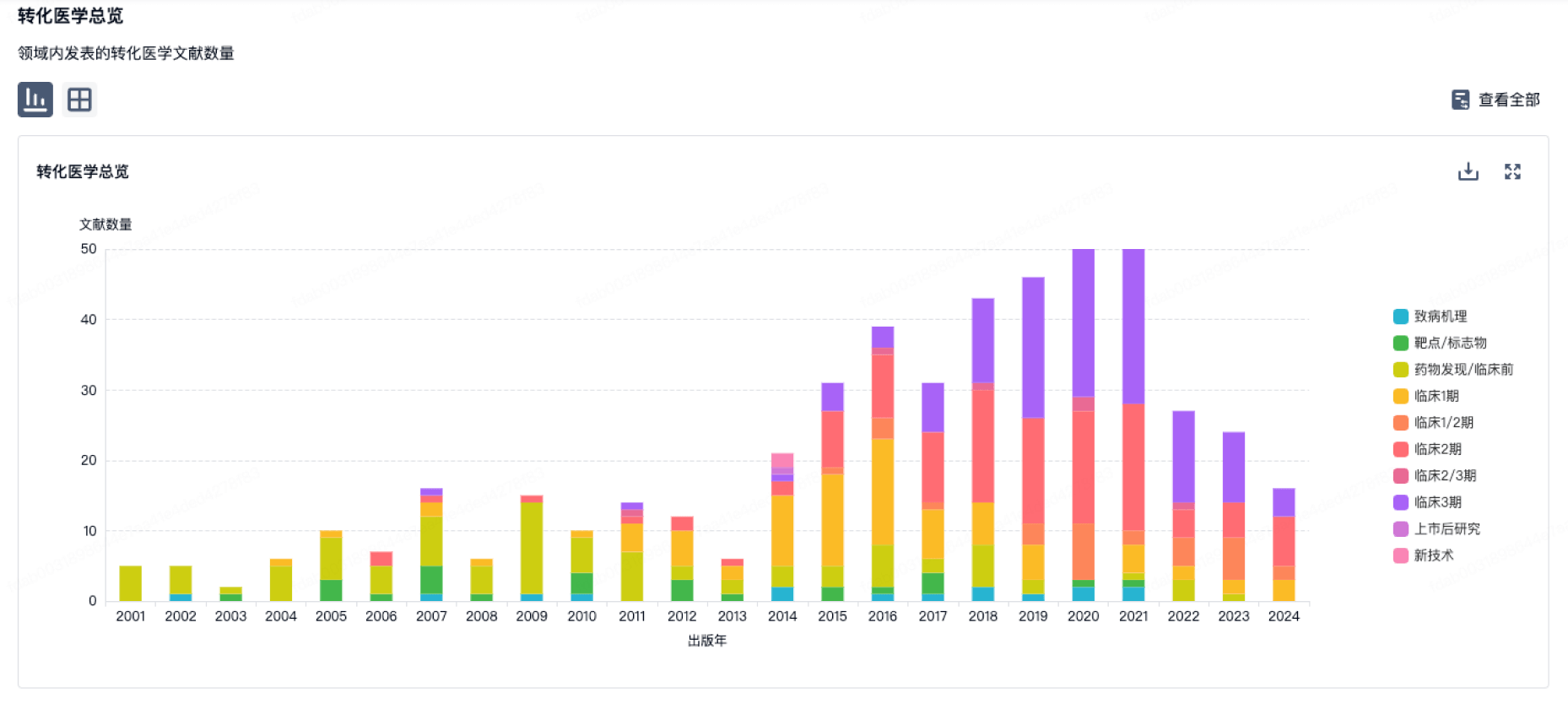
100 项与 上海博沃生物科技有限公司 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年07月01日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
药物发现
3
3
临床前
临床1期
1
3
其他
登录后查看更多信息
当前项目
| 药物(靶点) | 适应症 | 全球最高研发状态 |
|---|---|---|
下一代肺炎球菌结合疫苗(博沃生物) | 肺炎球菌感染 更多 | 临床前 |
BV-AdCoV-1 ( SARS-CoV-2 antigen ) | 新型冠状病毒感染 更多 | 临床前 |
广谱沙贝可疫苗(博沃) | 新型冠状病毒“贝塔”变异株感染 更多 | 临床前 |
五价减毒轮状病毒疫苗(博沃生物) | 轮状病毒感染 更多 | 药物发现 |
呼吸道合包病毒疫苗(博沃生物) | 呼吸道合胞体病毒感染 更多 | 药物发现 |
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用