预约演示
更新于:2025-08-08

Freenome Holdings, Inc.
更新于:2025-08-08
概览
关联
7
项与 Freenome Holdings, Inc. 相关的临床试验NCT06122077
The PROACT LUNG Study: A Prospective, Observational,Clinical Validation Study of the Freenome Multiomics Blood Test for Lung Cancer Screening (FRNM-007)
The PROACT LUNG study is a prospective multi-center observational study to validate a blood-based test for the early detection of lung cancer by collecting blood samples from high-risk participants who will undergo a routine, standard-of-care screening Low-Dose Computed Tomography (LDCT).
开始日期2023-11-28 |
申办/合作机构 |
NCT05516927
The Sanderson Study: A Diagnostic Case-Control Study for the Development of Multiomics Blood Tests for Cancer Screening in a Real World Setting
This protocol is a prospective, case-control multi-center diagnostic study to assess the sensitivity and specificity of blood-based screening tests for the early detection of multiple cancers.
开始日期2022-09-06 |
申办/合作机构 |
NCT05534906
The Detection of Small Early Liver Cancer With Natural History Follow up
The SELINA study will recruit 200 patients with cirrhosis and small HCC and 50 patients with HCC but without cirrhosis (most of whom are expected to have FLD). Blood, urine and liver tissue samples (where available) will be collected for laboratory analysis. In a subgroup of patients (N=80, around 64 patients with HCC with liver cirrhosis and around 16 patients with HCC without liver cirrhosis), additional magnetic resonance liver imaging will be performed. The findings of the SELINA study aim to identify biomarkers that can be used to detect liver cancer at the earliest possible time, something we expect will increase the survival rate of HCC.
开始日期2022-05-23 |
申办/合作机构  University of Oxford University of Oxford [+5] |
100 项与 Freenome Holdings, Inc. 相关的临床结果
登录后查看更多信息
0 项与 Freenome Holdings, Inc. 相关的专利(医药)
登录后查看更多信息
72
项与 Freenome Holdings, Inc. 相关的文献(医药)2025-11-01·EPILEPSY & BEHAVIOR
Social determinants of health (social risk) and epilepsy in older adults living in low-resource rural settings
Article
作者: Arias, Emilio E ; Mera, Robertino M ; Rumbea, Denisse A ; Del Brutto, Oscar H
BACKGROUND:
Information of the link between social risk and epilepsy in remote rural settings is limited. This study aims to assess this association in older adults enrolled in the Three Villages Study cohort.
METHODS:
Following a population-based cross-sectional design, older adults living in rural Ecuador underwent social risk determinations based on social determinants of health components from the Gijon's Social-Familial Evaluation Scale (SFES) together with clinical interviews to determine epilepsy history. Both unadjusted and multivariate logistic regression models were fitted to assess the association between the total Gijon's SFES and each of its components and epilepsy (dependent variable).
RESULTS:
The study included 682 individuals aged ≥ 60 years (mean age: 68 ± 7.3 years; 55 % women). The mean Gijon's SFES score was 10.1 ± 3.1 points, and the crude prevalence of epilepsy was 35.1 per 1,000 population. In unadjusted analysis participants in the highest tertile of social risk had significantly higher odds of having epilepsy compared to those in the lowest tertile (OR: 5.37; 95 % C.I.: 1.73 - 16.7). This association persisted when age, sex, and level of education were added to the model. Analysis of individual components of the Gijon's SFES showed that only social relationships and support networks were significantly associated with epilepsy.
CONCLUSION:
Study results indicate a link between high social risk and epilepsy. The direction of this association remains unclear, but a bidirectional relationship between both variables is likely. Adopting stronger community networks and social support systems could help mitigate epilepsy burden in low-resource settings.
2025-07-01·JAMA-JOURNAL OF THE AMERICAN MEDICAL ASSOCIATION
Clinical Validation of a Circulating Tumor DNA–Based Blood Test to Screen for Colorectal Cancer
Article
作者: Lee, Lilian C. ; Shaukat, Aasma ; Putcha, Girish ; Robertson, Douglas J. ; Gupta, Samir ; Burke, Carol A. ; Levin, Theodore R. ; Meng, Zhen ; Chan, Andrew T. ; Xu, Chuanbo ; Schoen, Robert E. ; Liu, Julia J. ; Grady, William M. ; Liang, Peter S. ; Ladabaum, Uri ; Lin, C. Jimmy ; Baldo, Lance ; Piscitello, Andrew ; Katona, Bryson W. ; Sun, Chung-Kai
Importance:
Colorectal cancer screening is widely recommended but underused. Blood-based screening offers the potential for higher adherence compared with endoscopy or stool-based testing but must first be clinically validated in a screening population.
Objective:
To evaluate the clinical performance of an investigational blood-based circulating tumor DNA test for colorectal cancer detection in an average-risk population using colonoscopy with histopathology as the reference method.
Design, Setting, and Participants:
Prospective, multicenter, cross-sectional observational study enrolling participants between May 2020 and April 2022 who were asymptomatic adults aged 45 to 85 years, at average risk of colorectal cancer, and willing to undergo a standard-of-care screening colonoscopy. Participants, staff, and pathologists were blinded to blood test results, and laboratory testing was performed blinded to colonoscopy findings. The study was conducted at 201 centers across 49 US states and the United Arab Emirates. Site-based and mobile phlebotomy were used for blood collection.
Exposures:
Participants were required to complete a screening colonoscopy after blood collection.
Main Outcomes and Measures:
The primary end points were sensitivity for colorectal cancer, specificity for advanced colorectal neoplasia (colorectal cancer or advanced precancerous lesions), negative predictive value for advanced colorectal neoplasia, and positive predictive value for advanced colorectal neoplasia. The secondary end point was sensitivity for advanced precancerous lesions.
Results:
The median age of participants in the evaluable cohort (n = 27 010) was 57.0 years, and 55.8% were women. Sensitivity for colorectal cancer was 79.2% (57/72; 95% CI, 68.4%-86.9%) and specificity for advanced colorectal neoplasia was 91.5% (22 306/24 371; 95% CI, 91.2%-91.9%). The negative predictive value for advanced colorectal neoplasia was 90.8% (22 306/24 567; 95% CI, 90.7%-90.9%) and the positive predictive value for advanced colorectal neoplasia was 15.5% (378/2443; 95% CI, 14.2%-16.8%). All primary end points met prespecified acceptance criteria. The sensitivity for advanced precancerous lesions was 12.5% (321/2567; 95% CI, 11.3%-13.8%), which did not meet the prespecified acceptance criterion.
Conclusions and Relevance:
In an average-risk colorectal cancer screening population, a blood-based test demonstrated acceptable accuracy for colorectal cancer detection, but detection of advanced precancerous lesions remains a challenge, and ongoing efforts are needed to improve test sensitivity.
Trial Registration:
ClinicalTrials.gov Identifier: NCT04369053
2025-07-01·Journal of Primary Care and Community Health
Prevalence of hypertriglyceridemic-waist phenotype and its association with type 2 diabetes mellitus among middle-aged and older adults of Amerindian ancestry
Article
作者: Del Brutto, Oscar H. ; Mera, Robertino M. ; Arias, Emilio E. ; Rumbea, Denisse A. ; Arriaga, Kleber
Background::
The hypertriglyceridemic-waist phenotype (HTWP), defined by concurrent hypertriglyceridemia and increased waist circumference, is a recognized marker of metabolic and cardiovascular risk. While extensively studied across populations, data on Amerindian communities remain scarce. This study examines HTWP prevalence and its association with type 2 diabetes mellitus in middle-aged and older adults of Amerindian ancestry in rural Ecuador.
Methods::
This population-based cross-sectional study was conducted in 3 ethnically homogeneous villages. Participants aged ≥40 years underwent standardized assessments, including structured interviews and fasting blood tests. HTWP was defined using serum triglyceride levels ≥150 mg/dL together with increased waist circumference determined by 2 criteria: Amerindian-specific (men ≥ 89 cm, women ≥83 cm) and NCEP-ATP III (men ≥102 cm, women ≥88 cm). Logistic regression models assessed associations between HTWP and diabetes indicators, adjusting for demographics and cardiovascular risk factors.
Results::
Among 1354 participants, HTWP prevalence was 47% by Amerindian-specific, and 30% using NCEP-ATP III criteria. Hypertriglyceridemia was frequent (55%), particularly in men. In multivariate models, HTWP was associated with fasting glucose ≥126 mg/dL under both Amerindian-specific (OR 1.32, 95% CI 1.02-1.71) and NCEP-ATP III (OR 1.50, 95% CI 1.12-2.01) criteria. When HTWP components were separately included in the models, only hypertriglyceridemia remained significantly associated with diabetes risk. No significant association was observed between HTWP and HbA1c levels.
Conclusion::
HTWP prevalence is high in this population. Hypertriglyceridemia drives diabetes risk more than waist circumference. Findings underscore the need for ethnicity-specific cardiovascular risk assessments and targeted health interventions for indigenous communities.
106
项与 Freenome Holdings, Inc. 相关的新闻(医药)2025-08-06
– Deal worth up to $885 million based on achievement of certain regulatory and screening guideline milestones –
– Final module for the first version of Freenome's colorectal cancer test has been submitted to FDA; approval and commercial launch anticipated in 2026 –
– Availability of real-world patient and multimodal molecular data will feed AI/ML models to improve Freenome's platform –
BRISBANE, Calif., Aug. 6, 2025 /PRNewswire/ -- Freenome, a biotechnology company pioneering an early cancer detection platform, today announced an exclusive license agreement with Exact Sciences to advance the commercialization of its colorectal (CRC) blood-based screening test,* including the U.S. commercial rights and the underlying technology. Freenome retains the rights for its CRC blood test when it is ordered in combination with additional cancer screening tests, including for lung and more than 10 other initial cancer indications the company is pursuing.
Exact Sciences will accelerate market adoption of the CRC blood test by leveraging its commercial infrastructure to streamline access to nearly 400 health systems with EMR integration; more than 865 in-network payers; more than 260,000 ordering physicians; and relationships with millions of patients who have been prescribed a Cologuard® test. Freenome will initially lead test processing, analysis and return of results while continuing to work with healthcare organizations to identify patients who are eligible for multiple tests.
"We are excited to enter into this agreement with Exact Sciences, which represents a pivotal moment in our mission to detect cancer in its earliest, most treatable stages," said Aaron Elliott, Ph.D., chief executive officer of Freenome. "With this agreement, our CRC blood test – which we believe is best in class, if approved – will be available much sooner to millions of patients. Additionally, Freenome will be able to augment our commercial reach and integration into primary care workflows, as we advance the development of our multiomic early cancer detection platform with the additional capital."
"This agreement in blood-based cancer screening accelerates our ability to bring new solutions to market," said Kevin Conroy, chairman and chief executive officer of Exact Sciences Corp. "As an additional option to Cologuard Plus™, this enhances our reach to unscreened patients. By integrating with our ExactNexus™ technology platform and commercial infrastructure, we're positioned to scale quickly and drive meaningful impact."
The terms include: an upfront payment of $75 million; $200 million in milestone payments associated with FDA first-line approval of the CRC blood test and a future test version; $500 million if the test is rated as an A or B test in the United States Preventive Services Taskforce (USPSTF) guidelines; royalties on test sales (expected to ramp to 10% once gross margins hit certain targets); $20 million in funding for joint R&D development expenses leveraging the technology for three years; and an equity investment of $50 million. Freenome will also have access to all multimodal data from patients to power future AI/ML models across multiple cancer indications.
"Population-level CRC screening is the gateway for our broader vision of personalized early cancer detection and, ultimately, disease detection beyond cancer," said Riley Ennis, Freenome's co-founder and chief product officer. "This partnership gives us the commercial scale and real-world data we need to improve our existing tests and accelerate development of the 10-plus other cancer types in our pipeline, and then expand from there."
CRC Screening Test Performance
In U.S.-census-adjusted data from Freenome's pivotal PREEMPT CRC® Study involving 48,995 average-risk adults, the company's CRC screening test detected 81.1% of CRC – including 63.5% at stage 1 – and 13.7% of advanced precancerous lesions (APL), with a specificity of 90.4%.1
As part of its test versioning strategy, Freenome has undertaken a comprehensive upgrade of the assay, automation and algorithm to develop an improved version of its CRC test. In a head-to-head study of the two versions on a primarily prospectively collected independent cohort, this "next-generation" version has shown even stronger performance in CRC and APL detection. The study included an average-risk, pre-colonoscopy arm to mirror the intended use population of PREEMPT CRC. Detailed data will be presented at an upcoming scientific meeting. Freenome is in active discussions with the FDA and plans to submit a supplemental premarket approval application for the next-generation test, following the approval of the first version.
CRC is the world's second deadliest cancer, with more than 50,000 deaths per year in the U.S. alone.2,3 When CRC is detected early, the survival rate is over 90%, yet more than 40% of screening-eligible adults are not up to date with current screening guidelines.4 The availability of less invasive, more accessible screening options – such as blood-based tests – has been shown to increase screening rates.5
Freenome and Early Cancer Detection
To pursue its vision of personalized early cancer detection, Freenome developed a multiomics platform that analyzes genomic, epigenomic, and proteomic biomarkers and applies AI/ML-based models to detect cancer-specific signals in the bloodstream, including those derived from circulating tumor DNA (ctDNA). The company's CRC screening blood test was built on this platform and identifies specific methylation signatures in ctDNA at single-base resolution.
Beyond CRC R&D and progressing its lung cancer laboratory-developed test toward an anticipated launch in 2026, Freenome will focus on developing its "personalized cancer early detection" program. The company's goal is to create a common lab platform with custom panels and classifiers to offer multiple tests to an individual, based on risk profiles and guideline eligibility. Targeting high-, elevated-, and average-risk populations based on the unmet need allows the tests to be optimized for higher sensitivity at clinically acceptable specificity.
About Freenome
Freenome is breaking barriers to early cancer detection with a suite of blood-based tests built on its multiomics platform. The company recognizes that no single technology can identify every cancer due to the disease's inherent heterogeneity. Freenome's multimodal approach combines molecular biology and assays with computational biology, machine learning and multiple data types to tune into cancer's subtlest cues, even at the earliest stages of the disease.
With the convenience of a standard blood draw, Freenome aims to empower everyone to access recommended cancer screenings. The company is partnering with healthcare organizations and population health decision-makers to integrate its technology and software platform, making cancer detection easier and more accessible. Freenome is headquartered in Brisbane, California. Find out more at and visit us on LinkedIn.
*Freenome's colorectal cancer blood-based screening test is referred to as "SimpleScreen™ CRC Colorectal Cancer screening blood test" in the company's Premarket Approval Application to the U.S. Food and Drug Administration.
References
Shaukat A, Burke CA, Chan AT, et al. Clinical validation of a circulating tumor DNA–based blood test to screen for colorectal cancer. JAMA. 2025;334(1):56-63. doi:10.1001/jama.2025.7515
National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer stat facts: common cancer sites. Accessed Aug. 6, 2025.
World Health Organization. Cancer. Published Feb. 3, 2025. Accessed Aug. 6, 2025.
Survival rates for colorectal cancer. American Cancer Society. Accessed Aug. 6, 2025.
Liang PS, Zaman A, Kaminsky A, et al. Clin Gastroenterol Hepatol. 2023 Apr 8;21(11):2951–2957.e2. doi: 10.1016/j.cgh.2023.03.036
SOURCE Freenome Holdings, Inc.
WANT YOUR COMPANY'S NEWS FEATURED ON PRNEWSWIRE.COM?
440k+
Newsrooms &
Influencers
9k+
Digital Media
Outlets
270k+
Journalists
Opted In
GET STARTED
引进/卖出临床结果
2025-06-02
– PREEMPT CRC, the largest prospective study of its kind, met all primary efficacy endpoints and surpassed CMS coverage requirements for sensitivity and specificity in the intended use population –
– FDA premarket approval submission is underway, with completion anticipated mid-2025 –
BRISBANE, Calif., June 2, 2025 /PRNewswire/ -- Freenome, a biotechnology company pioneering an early cancer detection platform, today announced the publication of detailed results from the pivotal PREEMPT CRC study in JAMA.1 The publication presents findings from the largest prospective study of a blood-based screening test for colorectal cancer (CRC), involving 48,995 average-risk adults aged 45 to 85 who underwent a routine colonoscopy following a blood draw.
Based on data from the 27,010 eligible participants who enrolled consecutively in the study after a predetermined cut-off date, the test met all prespecified primary acceptance criteria. The publication also includes a pre-specified analysis that weighted test performance to match the sex and age distribution of the U.S. population, a method used by the U.S. Food and Drug Administration (FDA) for other CRC screening products.2
"Despite clear guidelines, many communities still face barriers that lead to fewer people getting screened for colorectal cancer," said Aasma Shaukat, M.D., M.P.H., professor of medicine at NYU Grossman School of Medicine and a co-lead principal investigator on the PREEMPT CRC study. "The study's rigor and scale provide confidence in the test's performance. Its high sensitivity means it can detect most cancers, while high specificity helps avoid false alarms. Both are critical to making screening effective and efficient, as well as easier for patients and providers."
More than two out of five U.S. adults of screening age are not current with recommended CRC screening. Barriers such as discomfort, preparation or access often lead to low adoption rates. Importantly, among adults aged 45–49, who are newly recommended for screening and of whom only 20% are up to date, the test showed 100% sensitivity for CRC and 94.8% specificity for ACN in the primary analysis.
"By enrolling a diverse, average-risk population and analyzing samples consecutively, the PREEMPT study mirrored real-world use," said Aaron Elliott, Ph.D., chief executive officer at Freenome. "The test showed strong performance in Stage I cancer and high-risk pre-cancers and among younger adults, compared to FDA-approved CRC blood tests. This highlights the potential of the Freenome test to expand adherence to CRC screening recommendations and detect cancer earlier."
PREEMPT CRC was conducted at more than 200 clinical sites using a hybrid recruitment strategy with the end goal of increasing trial accessibility and diversity. This approach made participation possible from every state in the continental U.S. More than 11% of participants identified as Black or African American, and over 11% as Hispanic or Latino.
"We need more tools that meet people where they are, which includes offering noninvasive screening options that are simple to complete," Dr. Shaukat added. "This test has the potential to increase screening uptake, especially among people who might otherwise delay or avoid screening."
To pursue its vision of personalized early cancer detection, Freenome developed a multiomics platform that analyzes genomic, epigenomic, and proteomic biomarkers to detect cancer-specific signals in the bloodstream, including those derived from circulating tumor DNA (ctDNA). The company's CRC screening blood test was built on this platform and applies an AI/ML-based model to detect specific methylation signatures in ctDNA at a base level. Designed for use with a standard blood draw, the test integrates into routine clinical workflows, leveraging Freenome's software.
The company is advancing the premarket approval submission for its CRC screening blood test to the FDA, with all modules expected to be completed in mid-2025. Freenome also continues to advance a test versioning strategy focused on assay and algorithm improvements to enhance CRC and APL detection, as well as pursue expansion into additional indications, including lung cancer.
The full manuscript is now available online in JAMA. To access the paper, visit .
About PREEMPT CRC
PREEMPT CRC (NCT04369053) was a prospective, registrational clinical study designed to validate Freenome's blood test for the early detection of colorectal cancer (CRC) among average-risk adults. Initiated in 2020, the study was conducted at more than 200 sites and enrolled 48,995 asymptomatic, average-risk participants between the ages of 45 and 85 scheduled to undergo a screening colonoscopy. Freenome and the U.S. Food and Drug Administration agreed upon a predetermined cut-off date for the period most affected by the COVID-19 pandemic. The study analyzed the 27,010 participants who enrolled consecutively or after the predetermined cut-off date.
The study leveraged a novel hybrid model involving virtual and traditional recruitment methods to reach underserved communities and ensure a representative population. The decentralized recruitment strategy underscores Freenome's commitment to promoting equity and diversity in clinical studies, ensuring its tests are designed for everyone.
Freenome's partners in PREEMPT CRC included the Colorectal Cancer Alliance, Dia de la Mujer Latina, the Intercultural Center for Health Research and Wellness, and historically Black colleges and universities (HBCUs), including Morehouse School of Medicine. Freenome also worked with CVS Health Clinical Trial Services to help drive study enrollment through coordinated communication efforts targeting patients with already scheduled colonoscopies.
About Freenome
Freenome is breaking barriers to early cancer detection with a suite of blood tests built on its multiomics platform. The company recognizes that no single technology can identify every cancer due to the disease's inherent heterogeneity. Freenome's multimodal approach combines molecular biology and assays with computational biology, machine learning and multiple data types to tune into cancer's subtlest cues, even at the earliest stages of the disease.
With the convenience of a standard blood draw, Freenome aims to empower everyone to access recommended cancer screenings. The company is partnering with healthcare organizations and population health decision-makers to integrate its technology and software platform, making cancer detection easier and more accessible. Freenome is headquartered in Brisbane, California. Find out more at and visit us on LinkedIn.
Forward-Looking Statements
This press release contains forward-looking statements within the meaning of federal securities laws, including statements regarding the PREEMPT CRC, the use of information from that study, and Freenome's plans and expectations regarding its FDA submission, which involve risks and uncertainties that could cause the actual results to differ materially from the anticipated results and expectations expressed in these forward-looking statements. These statements are based on current expectations, forecasts, and assumptions, and actual outcomes and results could differ materially from these statements due to a number of factors. The forward-looking statements in this press release are based on information available to Freenome as of the date hereof, and Freenome disclaims any obligation to update any forward-looking statements provided to reflect any change in its expectations or any change in events, conditions, or circumstances on which any such statement is based, except as required by law. These forward-looking statements should not be relied upon as representing Freenome's views as of any date subsequent to the date of this press release.
References
1. Shaukat A, Burke CA, Chan AT, et al. Clinical Validation of a Circulating Tumor DNA-Based Blood Test to Screen for Colorectal Cancer. JAMA. Published online June 2, 2025. doi: 10.1001/jama.2025.7515.
2. U.S. Food and Drug Administration. Summary of safety and effectiveness data [Guardant Shield]. Published July 26, 2024. Accessed May 27, 2025.
3. Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA: A Cancer J for Clinicians. 2023;73(3):233-254. doi:10.3322/caac.21772.
4. Piscitello A, Edwards DK. Estimating the screening-eligible population size, ages 45–74, at average risk to develop colorectal cancer in the United States. Cancer Prevention Research. 2020;13(5):443-448. doi:10.1158/1940-6207.CAPR-19-0527.
SOURCE Freenome Holdings, Inc.
WANT YOUR COMPANY'S NEWS FEATURED ON PRNEWSWIRE.COM?
440k+
Newsrooms &
Influencers
9k+
Digital Media
Outlets
270k+
Journalists
Opted In
GET STARTED
临床结果
2025-05-21
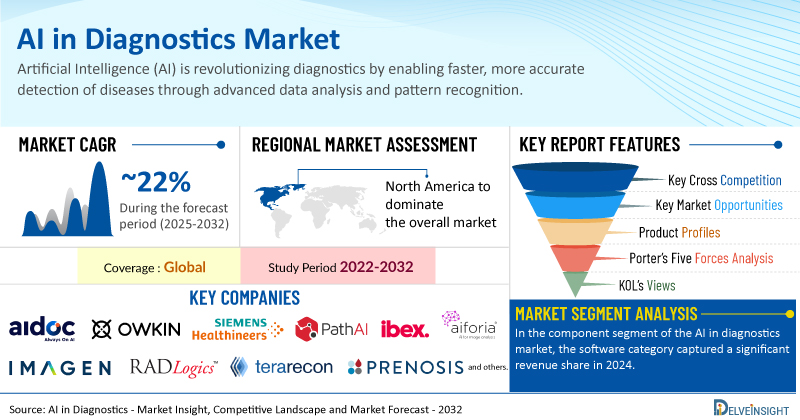
Global AI in Diagnostics Market to Register Stunning Growth at a CAGR of ~22% by 2032 | DelveInsight
The increasing incidence of infectious and chronic illnesses is boosting the need for early and precise diagnosis, with AI playing a crucial role by improving detection and enabling quicker clinical decisions. At the same time, the widespread adoption of digital health and imaging technologies creates a solid base for integrating AI into current healthcare systems, enhancing both efficiency and accuracy. Furthermore, ongoing product innovation by major global players, driven by progress in machine learning and growing regulatory backing, is accelerating the availability of AI-based diagnostic solutions.
New York, USA, May 21, 2025 (GLOBE NEWSWIRE) --
Global AI in Diagnostics Market to Register Stunning Growth at a CAGR of ~22% by 2032 | DelveInsight
The increasing incidence of infectious and chronic illnesses is boosting the need for early and precise diagnosis, with AI playing a crucial role by improving detection and enabling quicker clinical decisions. At the same time, the widespread adoption of digital health and imaging technologies creates a solid base for integrating AI into current healthcare systems, enhancing both efficiency and accuracy. Furthermore, ongoing product innovation by major global players, driven by progress in machine learning and growing regulatory backing, is accelerating the availability of AI-based diagnostic solutions.
DelveInsight’s
AI in Diagnostics Market Insights
report provides the current and forecast market analysis, individual leading AI in diagnostics companies’ market shares, challenges, AI in diagnostics market drivers, barriers, trends, and key market AI in diagnostics companies in the market.
Key Takeaways from the AI in Diagnostics Market Report
As per DelveInsight estimates, North America is anticipated to dominate the global AI in diagnostics market during the forecast period.
In the component segment of the AI in diagnostics market, the software category captured a significant revenue share in 2024.
Notable AI in diagnostics companies such as
Aidoc, Owkin, Inc., Siemens Healthineers, PathAI, Ibex, Owkin, Inc., Imagen Technologies, Aiforia, RADLogics, Terarecon, Inc., Prenosis, Inc., Ibex, Google LLC, GE HealthCare, DreaMed, Riverain Technologies, Terarecon, Inc., Aiforia, RADLogics,
and several others are currently operating in the AI in diagnostics market.
In
September 2024, Ibex Medical Analytics (Ibex)
, a leader in AI-powered cancer diagnostics, introduced the latest advancements in its innovative product platform, Ibex-AI. These new features, developed in collaboration with expert pathologists worldwide who actively use the platform in routine clinical practice, highlight Ibex's ongoing commitment to creating cutting-edge diagnostic tools tailored to the needs of healthcare professionals on the frontlines of patient care.
In
October 2023, Lucida Medical Ltd
announced that it received Class IIb CE certification for its AI-powered prostate cancer detection software, Prostate IntelligenceTM (PiTM). Leveraging advanced AI technology, PiTM analyzes MRI scans and is seamlessly integrated into the radiologist's workflow.
In
March 2023, Qritive
launched its latest AI-driven innovation, QAi Prostate, designed specifically for prostate cancer diagnosis. Utilizing cutting-edge machine learning algorithms, QAi Prostate analyzes whole slide images of prostate core needle biopsies with high precision.
To read more about the latest highlights related to the AI in diagnostics market, get a snapshot of the key highlights entailed in the
Global AI in Diagnostics Market Report
AI in Diagnostics Overview
Artificial Intelligence (AI) is revolutionizing diagnostics by enabling faster, more accurate detection of diseases through advanced data analysis and pattern recognition. AI-powered systems can analyze medical images such as X-rays, MRIs, and CT scans with high precision, often matching or exceeding the performance of human radiologists in identifying abnormalities like tumors, fractures, or infections. By processing vast amounts of data from electronic health records, lab results, and genomics, AI algorithms can also uncover hidden correlations, assist in early disease prediction, and support personalized treatment plans.
Moreover, AI is streamlining workflows in diagnostic laboratories and reducing the burden on healthcare professionals. For instance, AI-driven tools can automate routine tasks such as blood sample analysis or pathology slide interpretation, allowing clinicians to focus on complex cases. In resource-constrained settings, AI has the potential to bridge the gap in healthcare access by providing decision support to less experienced practitioners. As AI continues to evolve, integrating real-time data and improving interpretability, it holds the promise of significantly enhancing diagnostic accuracy, speed, and overall patient outcomes.
AI in Diagnostics Market Insights
North America is expected to dominate the overall Artificial Intelligence (AI) in Diagnostics market over the forecast period, driven by advanced healthcare infrastructure, significant investments in AI technologies, and widespread adoption of digital health solutions. The United States, in particular, leads the region due to its early adoption of cutting-edge technologies, strong presence of leading AI and healthcare companies, and supportive regulatory frameworks.
The growing demand for early and accurate diagnosis, especially for chronic and complex diseases such as cancer and cardiovascular conditions, has propelled the integration of AI into diagnostic tools and platforms. Major healthcare institutions in the region are increasingly leveraging AI algorithms for imaging analysis, pathology, and predictive diagnostics, further accelerating market growth.
Moreover, robust government initiatives and funding programs supporting AI research in healthcare continue to fuel innovation and development. Strategic collaborations between technology firms and medical institutions are resulting in AI solutions with improved accuracy and efficiency, contributing to enhanced patient outcomes.
The presence of a tech-savvy population, favorable reimbursement policies for AI-enabled diagnostic procedures, and a well-established ecosystem of data-driven healthcare systems also contribute to North America’s leading position in this domain. As AI technologies mature and gain greater clinical acceptance, the region is expected to remain at the forefront of global advancements in AI-driven diagnostics.
To know more about why North America is leading the market growth in the AI in diagnostics market, get a snapshot of the
AI in Diagnostics Market Outlook
AI in Diagnostics Market Dynamics
The AI in diagnostics market has seen significant growth due to
technological advancements in machine learning (ML) and artificial intelligence (AI)
, allowing for the automation and enhancement of diagnostic processes. AI-powered diagnostic tools are revolutionizing areas such as imaging, pathology, and molecular diagnostics, enabling faster, more accurate results with reduced human error. This has become especially crucial in areas like
cancer detection
, where early identification can significantly impact treatment outcomes. AI algorithms, capable of analyzing vast datasets, can detect patterns and abnormalities in medical imaging that may be missed by the human eye, leading to earlier intervention.
The market dynamics are also being influenced by the
increasing demand for personalized healthcare
. As healthcare systems move towards precision medicine, AI in diagnostics plays a vital role by tailoring diagnostic tests and treatments based on individual genetic profiles. This is particularly significant in oncology, where molecular diagnostics are being enhanced by AI to offer more targeted therapeutic options. With this move toward
personalized diagnostics
, the role of AI in predicting disease outcomes and treatment responses continues to expand, driving the need for more sophisticated diagnostic tools.
Furthermore,
regulatory approvals and integration with healthcare systems
are key to AI’s success in diagnostics. Governments and healthcare bodies are gradually developing guidelines for AI in clinical settings, which is boosting investor confidence and enabling more widespread adoption. However, challenges such as
data privacy concerns, interoperability issues
, and the
need for large-scale validation studies
still pose barriers to the full-scale integration of AI diagnostics. As a result, partnerships between AI companies and traditional diagnostic firms are becoming more common to combine the strengths of AI technologies with the reliability and credibility of established healthcare systems.
The
growing prevalence of chronic diseases
and the
increasing burden on healthcare systems
globally are also driving the adoption of AI in diagnostics. AI can assist in managing large patient populations by streamlining workflows, reducing diagnostic errors, and providing healthcare professionals with better decision-making tools. The market is also seeing the
emergence of AI-driven platforms
that offer end-to-end diagnostic solutions, from initial patient consultation to final diagnosis, creating a more seamless healthcare experience. As AI technologies continue to mature, the market is expected to experience even greater expansion, making healthcare more accessible, efficient, and accurate.
Get a sneak peek at the AI in diagnostics market dynamics @
AI in Diagnostics Market Dynamic Analysis
Report Metrics
Details
Coverage
Global
Study Period
2022–2032
AI in Diagnostics Market CAGR
~22%
AI in Diagnostics Market Size by 2032
USD 8 Billion
Key AI in Diagnostics Companies
Aidoc, Owkin, Inc., Siemens Healthineers, PathAI, Ibex, Owkin, Inc., Imagen Technologies, Aiforia, RADLogics, Terarecon, Inc., Prenosis, Inc., Ibex, Google LLC, GE HealthCare, DreaMed, Riverain Technologies, Terarecon, Inc., Aiforia, RADLogics, among others
AI in Diagnostics Market Assessment
AI in Diagnostics Market Segmentation
AI in Diagnostics Market Segmentation By Component:
Hardware/software, and Services
AI in Diagnostics Market Segmentation By Application:
Infectious Disease Diagnostics, Radiology, Oncology, Cardiology, and Others
AI in Diagnostics Market Segmentation By Technology:
Machine Learning, Natural Language Processing, and Others
AI in Diagnostics Market Segmentation By Geography
: North America, Europe, Asia-Pacific, and Rest of World
Porter’s Five Forces Analysis, Product Profiles,
Case Studies, KOL’s Views, Analyst’s View
Which MedTech key players in the AI in diagnostics market are set to emerge as the trendsetter explore @
AI in Diagnostics Companies
Table of Contents
1
AI in Diagnostics Market Report Introduction
2
AI in Diagnostics Market Executive Summary
3
Competitive Landscape
4
Regulatory Analysis
5
AI in Diagnostics Market Key Factors Analysis
6
AI in Diagnostics Market Porter’s Five Forces Analysis
7
AI in Diagnostics Market Layout
8
AI in Diagnostics Market Company and Product Profiles
9
KOL Views
10
Project Approach
11
About DelveInsight
12
Disclaimer & Contact Us
Interested in knowing the AI in diagnostics market by 2032? Click to get a snapshot of the
AI in Diagnostics Market Trends
Related Reports
AI in Cancer Diagnostics Market
AI in Cancer Diagnostics Market Insights, Competitive Landscape, and Market Forecast – 2032
report deliver an in-depth understanding of the market trends, market drivers, market barriers, and key AI in cancer diagnostics companies, including
iCAD, Inc., ibex-ai, Roche Diagnostics, Kheiron Medical Technologies Limited, MVision AI Inc., Siemens Healthineers AG, GE HealthCare, NVIDIA Corporation, Digital Diagnostics Inc., IBM Corporation, Azra AI, ConcertAI, PathAI, Median Technologies, Paige AI Inc., Therapixel, Flatiron, Freenome Holdings Inc., Onc.AI, Sonrai Analytics,
among others.
Artificial Intelligence In Clinical Trials Market
Artificial Intelligence In Clinical Trials Market Insights, Competitive Landscape, and Market Forecast – 2032
report delivers an in-depth understanding of the market trends, market drivers, market barriers, and key AI in clinical trials companies, including
TEMPUS, NetraMark, ConcertAI, AiCure, Medpace, Inc., ICON plc, Charles River Laboratories, Dassault Systèmes, Oracle, Certara, Cytel Inc., Phesi, DeepHealth, Unlearn.ai, Inc., H1, TrialX, Suvoda LLC, Risklick, Lokavant, Research Solutions
, among others.
Artificial Intelligence In Drug Discovery Market
Artificial Intelligence In Drug Discovery Market Insights, Competitive Landscape, and Market Forecast – 2032
report delivers an in-depth understanding of the market trends, market drivers, market barriers, and key AI in drug discovery companies, including
IBM Corporation, Numedii Inc, Deep Genomics, NVIDIA Corporation, Atomwise Inc, Cloud Pharmaceuticals Inc, Alphabet Inc (DeepMind), Insilico Medicine, BenevolentAI, Exscientia, Cyclia, Valo Health, Owkin Inc, Verge Genomics, BioSymetrics
, among others.
Artificial Intelligence in Drug Commercialization Market
Artificial Intelligence in Drug Commercialization Market Insight, Competitive Landscape, and Market Forecast – 2032
report delivers an in-depth understanding of market trends, market drivers, market barriers, and key AI in drug commercialization companies, including
EVERSANA, Lyfegen, Syneos Health, McKinsey & Company, ICON plc., Clarivate., Thermo Fisher Scientific Inc., Viseven, ZS Associates, Cloud Pharmaceuticals Inc.,
among others.
Artificial Intelligence in Precision Medicine Market
Artificial Intelligence in Precision Medicine Market Insight, Competitive Landscape, and Market Forecast – 2032
report delivers an in-depth understanding of market trends, market drivers, market barriers, and key AI in precision medicine companies, including
TEMPUS, GE HealthCare, Qure.ai, Envisionit Deep AI (Pty) Ltd., Avicenna.AI, Aignostics, Inc., Proscia Inc., Ultivue, Inc., Prenosis, Inc., IBEX, Cleerly, Inc., Paige AI, Inc., Densitas® Inc., Photocure ASA, iCAD, Inc., Eko Health, Inc., Owkin, Inc, Massive Bio, Deep Bio Inc., Atomwise Inc.,
among others.
About DelveInsight
DelveInsight is a leading Business Consultant and Market Research firm focused exclusively on life sciences. It supports pharma companies by providing comprehensive end-to-end solutions to improve their performance.
Contact Us
Shruti Thakur
info@delveinsight.com
+14699457679
https://www.delveinsight.com/medical-devices
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability
for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this
article. If you have any complaints or copyright issues related to this article, kindly contact the author above.
100 项与 Freenome Holdings, Inc. 相关的药物交易
登录后查看更多信息
100 项与 Freenome Holdings, Inc. 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年11月06日管线快照
无数据报导
登录后保持更新
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
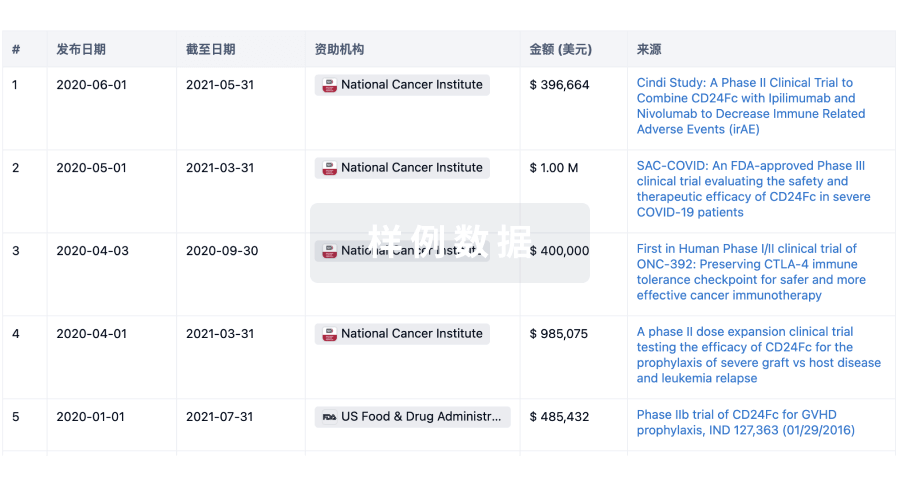
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
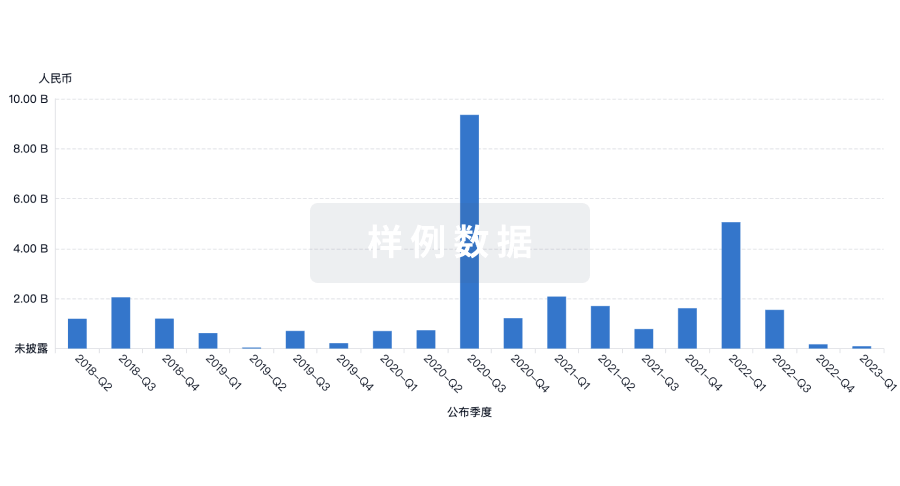
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用