预约演示
更新于:2025-09-09

Fujian Medical University
更新于:2025-09-09
概览
标签
肿瘤
皮肤和肌肉骨骼疾病
神经系统疾病
小分子化药
蛋白水解靶向嵌合体(PROTAC)
化学药
疾病领域得分
一眼洞穿机构专注的疾病领域
暂无数据
技术平台
公司药物应用最多的技术
暂无数据
靶点
公司最常开发的靶点
暂无数据
| 排名前五的药物类型 | 数量 |
|---|---|
| 小分子化药 | 19 |
| 蛋白水解靶向嵌合体(PROTAC) | 5 |
| 化学药 | 2 |
| 合成多肽 | 2 |
| 外泌体药物 | 2 |
关联
38
项与 福建医科大学 相关的药物作用机制 EDG6调节剂 [+3] |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2010-09-21 |
322
项与 福建医科大学 相关的临床试验ChiCTR2500106993
Research on the Influence Mechanism and Intervention Strategies of Co-parenting Conflicts on Parental Work Engagement in the Context of Intergenerational Caregiving
开始日期2025-08-01 |
申办/合作机构  福建医科大学 福建医科大学 [+1] |
ChiCTR2500106450
The Effects of Mirror Therapy Combined with Transcutaneous Electrical Nerve Stimulation on Limb Function in Stroke Patients and Its Neural Mechanisms: An fNIRS-Based Study
开始日期2025-07-28 |
申办/合作机构  福建医科大学 福建医科大学 [+1] |
NCT07081750
Development and Efficacy of a Dyadic Tailored Home-Based Activity Program for Mild-to-Moderate Dementia With Behavioral and Psychological Symptoms Post-Discharge: A Randomized Controlled Trial
This interventional study aims to evaluate the efficacy of a dyadic tailored home-based activity program in managing behavioral and psychological symptoms of dementia (BPSD) among home-dwelling people living with mild-to-moderate dementia post-discharge. The main question it aims to answer is:
Does the dyadic tailored home-based activity program significantly reduce BPSD in individuals with mild-to-moderate dementia after hospital discharge?
Researchers will compare people living with mild to moderate dementia who are receiving routine care to see if the dyadic tailored home-based activity program is effective for BPSD.
Participants will:
1. During hospitalization: People living with mild to moderate dementia and their caregivers receive health education on managing behavioral and psychological symptoms of dementia (BPSD); Participating activity sessions to assess individual preferences and functional activity capabilities, followed by co-designing a dyadic tailored home-based activity program.
2. After discharge: People living with mild to moderate dementia and their caregivers jointly receive a 12-week dyadic tailored home-based activity intervention at home.
Does the dyadic tailored home-based activity program significantly reduce BPSD in individuals with mild-to-moderate dementia after hospital discharge?
Researchers will compare people living with mild to moderate dementia who are receiving routine care to see if the dyadic tailored home-based activity program is effective for BPSD.
Participants will:
1. During hospitalization: People living with mild to moderate dementia and their caregivers receive health education on managing behavioral and psychological symptoms of dementia (BPSD); Participating activity sessions to assess individual preferences and functional activity capabilities, followed by co-designing a dyadic tailored home-based activity program.
2. After discharge: People living with mild to moderate dementia and their caregivers jointly receive a 12-week dyadic tailored home-based activity intervention at home.
开始日期2025-07-20 |
申办/合作机构 |
100 项与 福建医科大学 相关的临床结果
登录后查看更多信息
0 项与 福建医科大学 相关的专利(医药)
登录后查看更多信息
13,807
项与 福建医科大学 相关的文献(医药)2026-01-01·ANNALS OF VASCULAR SURGERY
Early Outcome of WeFlow-JAAA Off-the-Shelf Endograft in the Treatment of Juxtarenal/Pararenal Abdominal Aortic Aneurysms
Article
作者: Wang, Chunansheng ; Zhang, Lan ; Zuo, Jian ; Zhang, Hongpeng ; Zhuang, Hui ; Guo, Wei ; Dong, Honglin ; Zhang, Lei ; Chang, Guangqi ; Zhao, Jichun ; Li, Zhen ; Zhang, Xiaoming ; Gao, Jiangping ; Xin, Shijie ; Wang, Wei ; Guo, Pingfan ; Jiang, Jianjun ; Chen, Zhong ; Dai, Xiangchen ; Fu, Weiguo
BACKGROUND:
This study was performed to evaluate the early outcomes of the WeFlow-JAAA off-the-shelf (OTS) endograft system for the treatment of juxtarenal and pararenal abdominal aortic aneurysms (JR/PR-AAAs).
METHODS:
We conducted a prospective, multicenter study involving 115 patients diagnosed with JR/PR-AAAs. The inclusion criteria were a distance of ≥4 mm from the inferior edge of the superior mesenteric artery (SMA) to the aneurysm and a proximal landing zone angulation of <60°. All participants underwent endovascular repair using the WeFlow-JAAA system. The primary end points were clinical success and major adverse events (MAEs) within 30 days postprocedure. The secondary end points were all-cause mortality, secondary interventions, endoleaks, target vessel patency, and preservation of renal function during the first 30 days.
RESULTS:
The patients' mean age was 69.1 years, and 94.8% were male. The average maximum aneurysm diameter was 62.7 mm. The mean proximal neck length below the SMA was 20.3 ± 8.3 mm (range: 5-50 mm). Technical success was achieved in 99.1% of patients, with a target vessel patency rate of 99.4% within 30 days. One patient (0.9%) developed an early type I endoleak. MAEs occurred in 3.5% of patients, including 1 all-cause death due to cardiac arrest and three cases of minor ischemic stroke, all of which resolved. No significant aneurysm growth or stent migration was observed.
CONCLUSION:
The WeFlow-JAAA OTS endograft system demonstrated high technical success and low complication rates in the early treatment of JR/PR-AAAs. The occurrence of type II endoleaks and minor adverse events highlights areas for improvement. Future studies should focus on reducing type II endoleak risk and assessing long-term durability.
2026-01-01·TALANTA
A facile photoelectrochemical sensing platform for highly sensitive detection of dopamine secreted from nerve cells based on polydopamine sensitized TiO2 as photoelectric material
Article
作者: Yuan, Ya-Ni ; Li, Ji-Cheng ; Lei, Yun ; Liu, Ai-Lin
Dopamine (DA) is a catecholamine neurotransmitter in the brain, and changes in its concentration are closely related to neurological diseases. Thus, the development of a sensitive, cost-efficient and reliable method for DA monitoring has important application value. Herein, a photoelectrochemical (PEC) sensor based on TiO2 and polydopamine-modified TiO2 (PDA@TiO2) was constructed to achieve highly sensitive detection of DA secreted from living nerve cells. Upon DA introduction, PDA@TiO2 has a wide range of visible light absorption, promoting light absorption. The sensitization of PDA effectively improve the charge carrier transport efficiency, promote the photocatalytic activity of TiO2. The coating of PDA promotes its hydrophilic properties, effectively excluded the biofouling macromolecules and interference signals, ensuring sensor reproducibility. The result reflected that the current reponse increase of PDA@TiO2 is 3.5 times higher than the TiO2 after DA reaction due to the PDA@TiO2 can promote efficient carrier separation. The PDA@TiO2 sensor has superior sensitivity, a wider detection linear range and long-term stability. Notably, this PEC sensor is characterized by its simplicity in fabrication, low production cost, and portability. Under optimized experimental conditions, the PDA@TiO2 sensor showed a favorable linear response to DA using PBS incubated with pheochromocytoma (PC12) cells concentration ranging from 2 to 500 μM with a detection limit of 24.7 nM (S/N = 3). Meanwhile, the sensor was effectively applied to detect DA levels in PC12 cells. This work not only provides a new and efficient strategy for signal amplification, but also provides a promising candidate platform for sensitive detection of DA, offering a valuable tool for neuroscience and clinical diagnostics.
2026-01-01·CLINICA CHIMICA ACTA
Ensemble learning-driven hybrid prediction model for improved prenatal down’s syndrome screening: a comparative study with laboratory-based median equations
Article
作者: Wu, Xiaoshan ; Zhuang, Yingting ; Li, Yuqin ; Chen, Huan ; Xu, Minglin ; Hu, Liping ; Hong, Guolin ; Dai, Min ; Zhang, Zhimei
OBJECTIVE:
This study developed and validated a hybrid prediction model (HyPred) for prenatal Down's syndrome (DS) screening using ensemble learning(EL)-driven, including extreme gradient boosting, balanced random forest, and gradient boosting machine. Its performance was compared with laboratory-based median equations.
METHODS:
A retrospective analysis was conducted using 8,363 first-trimester samples (Nov 2019-Aug 2022) for training and 1,943 samples (Sep 2022-Jul 2023) for validation. HyPred was developed in R and compared with a laboratory-based median equation in terms of sensitivity, specificity, and other metrics.
RESULTS:
The lab-based median equation improved screening over the default equation. In the validation set, the EL model showed superior performance with higher accuracy, robustness, and adaptability. HyPred achieved an AUC of 0.97, surpassing the median equation in key metrics.
CONCLUSION:
The EL model offers improved accuracy and robustness for prenatal DS screening. Despite higher computational demands, modern tools facilitate its optimization, supporting precision medicine through more customized screening.
68
项与 福建医科大学 相关的新闻(医药)2025-08-29
2025年8月28日,在十五届中国—东北亚博览会上,韩国领先的生物技术公司JCBio与华大智造签署合作备忘录,共同启动韩国首个DCS Lab组学前沿实验室。双方将整合JCBio本土创新经验和华大智造创新测序技术平台,加速韩国在多组学研究领域的创新步伐,推动基因测序技术在精准医疗、临床诊断和AI数据分析等关键领域的深度应用与成果转化。
在十五届中国—东北亚博览会上,韩国生物技术公司JCBio与华大智造签署合作备忘录
JCBio首席执行官柳在燦(左)与华大智造东北亚区域总经理谭宏东博士(右)代表双方签约
打造一站式多组学平台
赋能韩国基因组学生态圈
成立于2005年的JCBio是一家韩国领先的生物科技公司,致力于运用前沿生物医学技术提升人类及灵长类动物的健康与寿命。面对多组学正在重塑医疗健康的时代机遇,此次共建的DCS Lab将成为赋能韩国基因组学生态圈的重要引擎。华大智造将通过其独有的DNBSEQ测序技术和多组学先进设备,为JCBio实验室构建一站式、端到端的多组学分析平台。该平台将显著提升研究人员应对复杂生物学挑战的精准度与效率,为韩国精准医疗及多组学研究成果的快速转化提供核心支撑。
JCBio首席执行官柳在燦表示:“为韩国科研人员提供高效、优质的生命核心工具是JCBio一以贯之的目标。我们非常荣幸能与华大智造合作,共建世界一流的多组学研究平台。依托华大智造的先进长读长及短读长测序技术,JCBio实验室将为韩国研究者提供解决复杂生物学难题的尖端测序工具,推动精准医疗与多组学研究的突破和发展。”
华大智造东北亚区域总经理谭宏东博士表示:“我们高度认可JCBio对科学卓越与创新的不懈追求。在此次合作中,华大智造的角色远不止于提供测序平台,更将与JCBio携手,将DCS Lab打造成为多组学发现中心,为韩国科研界输送推动前沿科学发展和改善人类健康的核心工具与创新能力。”
DCS Lab实验室赋能计划:
探索全球前沿生命科学边界
DCS Lab是华大智造于2023年推出的全球性实验室赋能计划,通过集成基因组学(DNA Omics,简称“D”)、细胞组学(Cell Omics,简称“C”)、时空组学(Spatial Omics,简称“S”)三大先进技术,为科研人员提供同时在DNA、细胞、组织等不同维度,对样本进行批量处理和成体系化的研究能力。该计划旨在助力全球顶级科研人员打造国际领先的、规模化、标准化的多组学前沿实验室,为生命科技行业开拓更多可能。
截至目前,DCS Lab已先后在上海脑科学与类脑研究中心、细胞生态海河实验室、新加坡国立癌症中心、北京大学现代农业研究院、福建医科大学、中国农业大学国家模式动物科学中心、合肥大健康研究院等地落成。此次与JCBio合作将DCS Lab首次引入韩国,再次彰显了华大智造以创新工具为核心、持续赋能全球生命科学前沿发现的坚定承诺。
截至2025年6月30日,华大智造业务遍及六大洲超过110多个国家和地区,在全球服务累计超过3,560个用户。未来,华大智造将将始终秉承“创新智造引领生命科技”的理念,通过持续的技术革新,为全球用户提供精准、高效、可靠的多组学工具平台及全流程支撑,携手全球伙伴开拓生命科学边界,共同拥抱“SEQ ALL”多组学融合新时代。
·
·
引进/卖出
2025-08-14
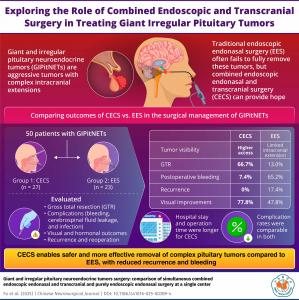
Two-team surgical approach removes more complex pituitary tumors, reduces bleeding risk, and lowers recurrence compared to traditional endonasal surgery
BEIJING, BEIJING, CHINA, August 14, 2025 /
EINPresswire.com
/ -- The pituitary gland, located at the base of the brain, secretes hormones that regulate vital body functions and control the activity of other hormone-secreting glands. Pituitary neuroendocrine tumors (PitNETs) are abnormal growths in this gland. In recent years, endoscopic endonasal surgery (EES), a minimally invasive technique, has become a widely used method for treating these tumors. In this approach, an endoscope is inserted through the nasal passages and sinuses—a route referred to as endonasal.
However, giant and irregular pituitary neuroendocrine tumors (GIPitNETs) pose a significantly greater challenge. These tumors are typically larger than 4 cm and often extend beyond the sella, the bony structure that houses the pituitary gland. They may grow upwards into the cranial cavity, the space within the skull that houses the brain. This extension can render the tumor inaccessible or invisible through the standard EES approach.
An alternative and innovative method used in such cases is the combined endoscopic endonasal and transcranial surgery (CECS). In this technique, one surgical team performs the EES approach while another team simultaneously carries out transcranial surgery, which involves accessing the tumor by creating small openings in the skull.
To analyze and compare the efficacy and complications of CECS and EES for GIPitNETs treatment, a team of researchers from China conducted a retrospective observational single-center cohort study. The study led by Dr. Changzhen Jiang and Dr. Xiaorong Yan from the Neurosurgery Research Institute, Fujian Medical University, China, was published in the
Chinese Neurosurgical Journal
and made available online on February 03, 2025. “We wanted to define the limitations and benefits of the two surgical procedures in the management of GIPitNETs,” says Dr. Jiang while explaining the aim of the study.
The research included 50 patients who underwent either EES or CECS between March 2018 and May 2023 at The First Affiliated Hospital of Fujian Medical University. All patients had tumors larger than 4 cm with significant intracranial extension. Endocrine tests were conducted before and after surgery to assess hormone levels. Magnetic resonance imaging (MRI) was used to evaluate tumor size and post-surgical outcomes. Researchers also analyzed hospital records for symptoms and complications, and ophthalmologists assessed visual function before and after surgery.
27 out of the 50 patients enrolled for this study were treated by CECS, and EES was performed on the remaining 23 patients. The researchers compared the obtained data using statistical analysis.
The results revealed a higher rate of gross total tumor removal (GTR) in the CECS group. GTR was achieved in 66% of patients in the CECS group, compared to just 13% in the EES group. Postoperative bleeding, a common and serious complication, was more prevalent in the EES group—65.2% compared to 7.4% in the CECS group. Additionally, all four cases of tumor recurrence due to residual tumor were reported in the EES group, suggesting that incomplete removal in EES may increase the risk of recurrence.
Interestingly, even though CECS achieved better tumor removal, visual outcomes were similar in both groups. “Partial tumor removal can also alleviate the pressure on optic nerves and lead to symptom relief. Thus, visual symptom improvement is possible after undergoing EES,” explains Dr. Yan.
However, CECS is not without its limitations. “The CECS technique requires a longer operation time and has greater surgical trauma with similar postoperative infection rates, compared to EES,” notes Dr. Jiang. Patients in the CECS group also had longer hospital stays. Despite the increased invasiveness of CECS, postoperative infection rates were comparable to those of the less invasive EES, indicating that CECS remains a safe option when performed carefully by experienced surgical teams.
Overall, the findings suggest that CECS may offer significant advantages in treating GIPitNETs, particularly when the tumor’s size or shape makes it inaccessible by EES alone. The improved GTR rate and lower complication rate point to CECS as a more effective approach for complex cases, despite the longer operation and recovery times.
The research team plans to further investigate the long-term efficacy and safety of CECS. They aim to conduct follow-up studies over extended periods and hope to analyze more patient data through large-scale multicenter collaborations. With continuous improvements in surgical techniques, approaches like CECS may help make the treatment of complex tumors like GIPitNETs safer and more successful.
***
Reference
Title of original paper: Giant and irregular pituitary neuroendocrine tumors surgery: comparison of simultaneous combined endoscopic endonasal and transcranial and purely endoscopic endonasal surgery at a single center
Journal: Chinese Neurosurgical Journal
DOI: 10.1186/s41016-025-00389-4
About Fujian Medical University
Fujian Medical University (FJMU), established in 1937, is a leading medical research university in China. It has over 20,000 full-time students and a strong faculty, more than half of whom hold doctorates or other advanced degrees. FJMU drives innovation in molecular medicine, oncology, and public health. The university hosts several national and provincial research platforms, including key laboratories and clinical trial centers. FJMU’s emphasis on translational medicine and interdisciplinary collaboration fosters cutting-edge discoveries. FJMU also participates in global research networks and offers English-medium postgraduate programs, attracting international scholars and supporting China’s broader scientific and medical advancement.
Website:
https://oec.fjmu.edu.cn/en/
About Dr. Xiorong Yan, from Fujian Medical University
Dr. Xiaorong Yan is a neurosurgeon at the Neurosurgery Research Institute, First Affiliated Hospital of Fujian Medical University in Fuzhou, China. She holds an MD in Neurosurgery from Shandong Medical University. Her work mainly focuses on endoscopic surgery for pituitary adenomas and other brain lesions. She co-led the recent comparison study of combined endoscopic endonasal‑transcranial surgery versus purely endonasal surgery for giant pituitary tumors, contributing to improved surgical strategies and patient outcomes. Dr. Yan has also contributed to other minimally invasive neurosurgical techniques, publishing clinical studies on spine and brain‑tumor procedures. She has over 20 published papers.
Funding information
This work was sponsored by Joint Funds for Innovation of Science and Technology, Fujian Province (No. 2021Y9089), University-Industry Research Joint Innovation Project of Science and Technology, Fujian Province (No. 2023Y4018), Fujian Province Finance Project (No. BPB-2022YXR), scientific research, Fujian Medical University (grant number 2022QH1098).
Yi Lu
Chinese Neurosurgical Journal
+861059978478 ext.
- luyi617@sina.cn
Legal Disclaimer:
EIN Presswire provides this news content "as is" without warranty of any kind. We do not accept any responsibility or liability
for the accuracy, content, images, videos, licenses, completeness, legality, or reliability of the information contained in this
article. If you have any complaints or copyright issues related to this article, kindly contact the author above.
临床结果
2025-08-13
·生物世界
撰文丨王聪
编辑丨王多鱼
排版丨水成文
急性胰腺炎(AP)与高死亡率相关,其特征是腺泡细胞(Acinar cell)死亡增加,消化酶过早释放和激活。在急性期,急性胰腺炎伴有“胞葬作用”(efferocytosis)增强,以清除凋亡细胞;Anxa1 蛋白对于胞葬作用至关重要,但其在急性胰腺炎中的作用仍不清楚。
2025 年 8 月 11 日,福建医科大学附属协和医院潘誉、黄鹤光,澳门科技大学尹成亮等在 Nature Nanotechnology 上发表了题为:Annexin A1 mRNA-loaded liposomes alleviate acute pancreatitis by suppressing STING pathway and promoting efferocytosis in macrophages 的研究论文。
该研究开发了一种新型 mRNA 疗法用于缓解急性胰腺炎,使用纳米脂质体递送 Anxa1 mRNA,通过抑制 STING 通路并促进巨噬细胞的胞葬作用,实现对急性胰腺炎的缓解。
急性胰腺炎(AP)的特征是腺泡细胞发生坏死性细胞死亡,导致胰腺坏死,同时释放与核损伤相关的分子模式、促炎介质和炎症趋化因子。在急性胰腺炎的急性期,巨噬细胞迅速清除被吞噬的凋亡细胞,这一过程被称为“胞葬作用”(efferocytosis),以防止出现不适当的炎症反应。胞葬作用是一种高度保守的过程,由多种吞噬配体触发,包括 Anxa1 蛋白。Anxa1 以钙依赖的方式与凋亡细胞表面的磷脂酰丝氨酸结合,并促进巨噬细胞对凋亡细胞的吞噬。近期的研究表明,STING 信号通路的激活与急性胰腺炎(AP)的发病机制有关。
在过去的几年里,基于 mRNA 的疗法已作为一种治疗策略崭露头角。然而,mRNA 稳定性差限制了其临床应用。为了解决这一问题,研究人员使用纳米载体进行 mRNA 递送,以提高 mRNA 的其稳定性和靶向能力,其中,纳米脂质体已被用于 siRNA 和 mRNA 的递送载体,但基于纳米脂质体的 mRNA 递送系统在临床应用中仍受到一定限制,部分原因在于其在血液循环过程中会被免疫系统清除。
在这项最新研究中,研究团队证实,Anxa1 蛋白的缺乏会消除胰腺巨噬细胞的胞葬作用,导致凋亡的腺泡细胞积聚和坏死。
在这一发现的基础上,研究团队进一步证明,负载 Anxa1 mRNA 的纳米脂质体能够通过抑制 cGAMP-cGAS-STING 通路,恢复巨噬细胞的胞葬作用,从而缓解急性胰腺炎(AP)的病理状况。
总的来说,这项研究揭示了 Anxa1 在急性胰腺炎期间巨噬细胞的胞葬作用中的关键功能,并展示了一种治疗急性胰腺炎的新型纳米技术手段,这可能对人类具有潜在的治疗价值。
论文链接:
https://www-nature-com.libproxy1.nus.edu.sg/articles/s41565-025-01979-0
设置星标,不错过精彩推文
开放转载
欢迎转发到朋友圈和微信群
微信加群
为促进前沿研究的传播和交流,我们组建了多个专业交流群,长按下方二维码,即可添加小编微信进群,由于申请人数较多,添加微信时请备注:学校/专业/姓名,如果是PI/教授,还请注明。
点在看,传递你的品味
信使RNAsiRNA寡核苷酸
100 项与 福建医科大学 相关的药物交易
登录后查看更多信息
100 项与 福建医科大学 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年10月31日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
药物发现
4
32
临床前
临床申请批准
1
1
临床1期
其他
7
登录后查看更多信息
当前项目
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
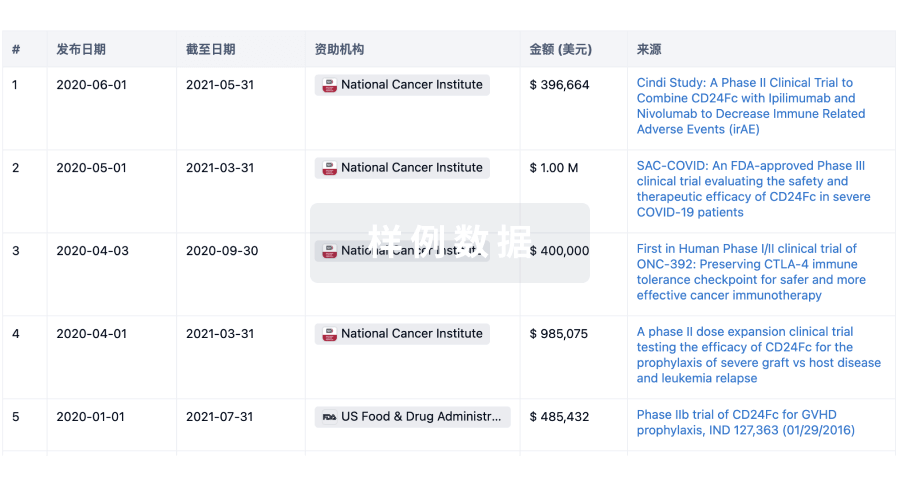
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
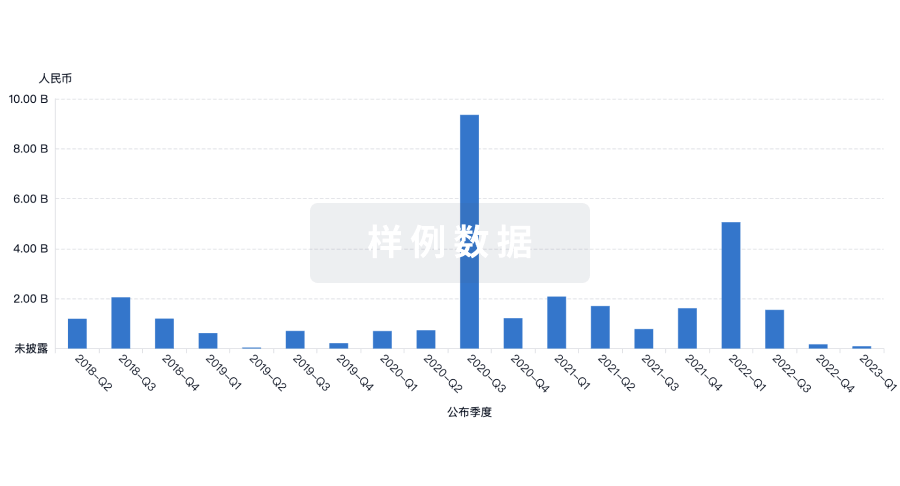
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

