预约演示
更新于:2025-05-07
Olsalazine Sodium
奥沙拉秦钠
更新于:2025-05-07
概要
基本信息
原研机构 |
非在研机构- |
权益机构- |
最高研发阶段批准上市 |
首次获批日期 美国 (1990-07-31), |
最高研发阶段(中国)批准上市 |
特殊审评- |
登录后查看时间轴
结构/序列
分子式C14H10N2Na2O6 |
InChIKeyWBFGLVPPGBVMLC-UHFFFAOYSA-N |
CAS号6054-98-4 |
查看全部结构式(2)
关联
2
项与 奥沙拉秦钠 相关的临床试验NCT02709824
Efficacy of an Anti-Discoloration System (ADS) to Reduce Dental Staining in a 0.12% Chlorhexidine-based Mouthwash
Dental staining is the most common local side effect of chlorhexidine (CHX) mouthwashes.
A prospective, cross-over, randomized clinical trial will be carried out in triple blind to compare the effect on dental staining of two 0.12% CHX mouthwashes: with or without anti-discoloration system (ADS). Twenty-two healthy subjects will be enrolled. Briefly, the study includes two sequential stages: in the first phase, the "immediate" objective and subjective evaluation of dental discoloration by spectroscopic and questionnaire-based analyses will be recorded, at baselines and after each 21-days-long cycle of mouthwash. A wash-out period between the cycles of 21 days will be performed.
In the second phase, a long-term (after 6 months) subjective evaluation of dental discoloration will be obtained by means of clinical photograph visual analysis and new questionnaires.
A prospective, cross-over, randomized clinical trial will be carried out in triple blind to compare the effect on dental staining of two 0.12% CHX mouthwashes: with or without anti-discoloration system (ADS). Twenty-two healthy subjects will be enrolled. Briefly, the study includes two sequential stages: in the first phase, the "immediate" objective and subjective evaluation of dental discoloration by spectroscopic and questionnaire-based analyses will be recorded, at baselines and after each 21-days-long cycle of mouthwash. A wash-out period between the cycles of 21 days will be performed.
In the second phase, a long-term (after 6 months) subjective evaluation of dental discoloration will be obtained by means of clinical photograph visual analysis and new questionnaires.
开始日期2011-12-01 |
申办/合作机构 |
NCT00004288
Phase II Pilot Study of Olsalazine for Ankylosing Spondylitis
OBJECTIVES:
I. Assess the safety and efficacy of olsalazine, a dimer of 5-aminosalicylic acid, in men with ankylosing spondylitis unresponsive to nonsteroidal anti-inflammatory drugs and physiotherapy.
I. Assess the safety and efficacy of olsalazine, a dimer of 5-aminosalicylic acid, in men with ankylosing spondylitis unresponsive to nonsteroidal anti-inflammatory drugs and physiotherapy.
开始日期1996-05-01 |
申办/合作机构 |
100 项与 奥沙拉秦钠 相关的临床结果
登录后查看更多信息
100 项与 奥沙拉秦钠 相关的转化医学
登录后查看更多信息
100 项与 奥沙拉秦钠 相关的专利(医药)
登录后查看更多信息
1,025
项与 奥沙拉秦钠 相关的文献(医药)2025-04-01·Journal of Advanced Research
Association of intestinal anti-inflammatory drug target genes with psychiatric Disorders: A Mendelian randomization study
Article
作者: Zhao, Guorui ; Kang, Zhewei ; Sun, Junyuan ; Sun, Yaoyao ; Liao, Yundan ; Feng, Xiaoyang ; Yue, Weihua ; Zhang, Yuyanan ; Lu, Zhe
2025-03-20·International Journal of Neuropsychopharmacology
Antidepressants in the treatment of bipolar depression: commentary
Review
作者: Vázquez, Gustavo H ; Baldessarini, Ross J
2025-03-01·Progress in Neuro-Psychopharmacology and Biological Psychiatry
A naturalistic retrospective evaluation of the utility of pharmacogenetic testing based on CYP2D6 e CYP2C19 profiling in antidepressants treatment in a cohort of patients with major depressive disorder
Article
作者: Squassina, Alessio ; Pinna, Marco ; Manchia, Mirko ; Contu, Martina ; Gennarelli, Massimo ; Paribello, Pasquale ; Congiu, Donatella ; d'Aversa, Federico Bernoni ; Severino, Giovanni ; Conversano, Claudio ; Del Zompo, Maria ; Pinna, Federica ; Carpiniello, Bernardo ; Pisanu, Claudia ; Minelli, Alessandra ; Meloni, Anna ; Mola, Francesco ; Carta, Andrea
2
项与 奥沙拉秦钠 相关的新闻(医药)2023-12-05
·今日头条
12月05日的《热心肠日报》,我们解读了 10 篇文献,关注:肠道免疫,菌群-免疫互作,多发性硬化,IBD,纳米医学,牙釉质,麸质敏感,1型糖尿病。
Cell子刊:STING进核激活AHR,促进共生菌和肠道稳态
Immunity——[32.4]
① 核定位的STING1可驱动转录因子芳烃受体(AHR)激活,来控制肠道微生物群组成和稳态;② 该功能独立于DNA感知和自噬,并与细胞质cGAS-STING1信号表现出竞争性抑制作用;③ STING1的环状二核苷酸结合结构域与AHR的N-末端结构域相互作用,但STING1介导的AHR转录激活需要额外的核伴侣,包括正向和负向调节蛋白;④ AHR配体可以改善野生型小鼠的结肠炎病理和微生态失调,但STING1的失活突变可消除这种保护作用。
【主编评语】
STING1(也称为STING)蛋白作为信号中枢,调控细胞对胞质内的异位DNA的免疫反应和自噬反应。Immunity近期发表的文章,发现STING1可定位到细胞核中,来驱动转录因子AHR的激活。STING1-AHR互作可保护小鼠抵抗肠道炎症和菌群失调。(@章台柳)
【原文信息】
Nuclear localization of STING1 competes with canonical signaling to activate AHR for commensal and intestinal homeostasis
2023-11-27, doi: 10.1016/j.immuni.2023.11.001
Cell子刊:肠菌如何塑造有抗炎作用的Th17
Immunity——[32.4]
① 通过探索共生微生物诱导肠道Th17细胞抗炎属性,表现为IL-10和共抑制受体表达,由转录因子c-MAF驱动;② 分节丝状细菌(SFB)等微生物特异性诱导的Th17细胞是异质的,起源于TCF1+肠道驻留的祖先Th17细胞群;③ SFB诱导Th17细胞抗炎转录程序与其他肠道Th17细胞不同,类似于耗竭和调节性CD4+ T细胞;④ 特异性删除Th17细胞c-MAF会导致其失去抗炎表型并增加促炎特性;⑤ TCF1+祖先Th17细胞向抗炎IL-10的转变会受到IL-10信号和肠道巨噬细胞影响。
【主编评语】
共生微生物诱导产生细胞因子的效应组织驻留CD4+ T细胞,但这些T细胞在粘膜稳态中的功能尚不清楚。近日,哥伦比亚大学研究人员在Immunity发表最新研究,展示Th17细胞的异质性和在肠道耐受中的专门调节功能,突显了特异性共生T细胞在维持稳态方面超越病原体控制的被低估的作用,对炎症和微生物组免疫互作产生影响,并为在炎症性肠病中治疗性地靶向SFB Th17细胞轴提供了理论基础,值得关注。(@九卿臣)
【原文信息】
Intestinal microbiota-specific Th17 cells possess regulatory properties and suppress effector T cells via c-MAF and IL-10
2023-11-30, doi: 10.1016/j.immuni.2023.11.003
国内团队Cell子刊:多发性硬化两性差异背后的肠道机制
Immunity——[32.4]
① 多发性硬化患者(MS)粪便苯乙胺含量显著高于健康对照且存在性别差异,基因敲除发现小鼠肠上皮细胞只有多巴胺受体D2时,可显著缓解MS疾病进展;② 在DRD2敲除的MS小鼠模型中,伴随着抗菌肽表达量显著减少和肠菌改变,且DRD2作用依赖于肠菌;③ 鉴定出47种只在雌鼠脊髓中有差异的代谢物,其中N-乙酰赖氨酸可显著抑制炎症反应,缓解自身免疫性脑脊髓炎发病;④ N-乙酰赖氨酸可显著降低MS相关小胶质细胞比例,提高增殖性和稳态小胶质细胞比例。
【主编评语】
多发性硬化症在发病率和临床特征上表现出强烈的女性倾向,其患者多为中青年女性。但目前已有的治疗药物多为对症治疗,选择品种有限且价格昂贵,无法得到根治,给家庭和社会带来巨大的负担。近日,中国科学院脑科学与智能技术卓越创新中心周嘉伟、中国科学院分子细胞科学卓越创新中心宋昕阳、中国科学院上海有机化学研究所生物与化学交叉研究中心朱正江、上海交通大学医学院附属瑞金医院神经内科陈晟及团队在Immunity发表最新研究,发现肠道上皮细胞多巴胺D2受体(IEC DRD2)过度激活可选择性地在雌性小鼠中改变肠道菌群的组成及其代谢物水平,从而促进多发性硬化的发病,值得关注。(@九卿臣)
【原文信息】
Intestinal epithelial dopamine receptor signaling drives sex-specific disease exacerbation in a mouse model of multiple scleROSis
2023-11-21, doi: 10.1016/j.immuni.2023.10.016
Nature子刊:新型纳米药物或可用于IBD持久治疗
Nature Biomedical Engineering——[28.1]
① 将磷酸胆碱(PC)修饰到纤维形成肽Q11,可自发聚集形成纳米纤维,免疫小鼠导致其产生针对小分子表位PC的天然抗体;② 这种抗PC的天然抗体具有抗感染和抗炎特性,因此,这种纳米纤维诱导高水平的抗PC抗体,在多个结肠炎小鼠模型中具有保护和治疗作用;③ 调整纳米纤维上的PC表位、调整外源性T细胞表位的相对价态以及添加佐剂CpG,都能控制纳米纤维引发的免疫反应的强度和类型。
【主编评语】
Nature Biomedical Engineering近期发表的文章,构建了新型纳米纤维,诱导小鼠产生抗磷酸胆碱的抗体,从而长久地缓解小鼠的结肠炎。这种纳米材料辅助诱导产生治疗性抗体,或可成为炎症性肠病持久治疗的一种方法。(@章台柳)
【原文信息】
Engaging natural antibody responses for the treatment of inflammatory bowel disease via phosphorylcholine-presenting nanofibres
2023-11-27, doi: 10.1038/s41551-023-01139-6
四川大学:奥沙拉秦纳米针与菊粉凝胶,双剑合璧治肠炎
Bioactive Materials——[18.9]
① 开发了一种基于奥沙拉秦(Olsa)的纳米针和微生物群调节菊粉凝胶的组合,其具有微米大小的针状结构及ROS清除和抗炎作用;② 纳米针和菊粉凝胶(Cu2(Olsa)/Gel)复合材料表现出大孔结构,可改善生物粘附性,口服后增强了结肠保留;③ 该复合材料通过抗炎和抗氧化疗法可有效下调促炎细胞因子水平并促进上皮屏障修复,从而显著缓解三种IBD动物模型中的结肠炎;④ Cu2(Olsa)/Gel处理会增加肠道菌群的多样性,降低变形菌门等病原菌的相对丰度。
【主编评语】
越来越多的数据证明,肠道菌群在肠道稳态和生态失调中起着重要作用,微生物群落代谢物的改变与IBD的发生和发展有关。但口服药物还受到胃肠道中复杂恶劣的理化环境的挑战,如酶、细菌、不同的pH值和加速的肠道蠕动。近日,四川大学何斌、蒲雨吉及团队在Bioactive Materials发表最新研究,开发了一种口服Cu2(Olsa) 纳米针菊粉凝胶复合物,用于治疗炎症性肠病,在小鼠中实测效果较好,或可用于重塑肠道稳态和治疗IBD,值得关注。(@九卿臣)
【原文信息】
An olsalazine nanoneedle-embedded inulin hydrogel reshapes intestinal homeostasis in inflammatory bowel disease
2023-11-08, doi: 10.1016/j.bioactmat.2023.10.028
克罗恩病全基因组测序研究,揭示种族混血对健康影响的新见解
Genome Medicine——[12.3]
① 纳入3418名非裔美国人的全基因组测序数据;② 另一项欧洲队列中发现的溃疡性结肠炎(CD)相关的25个罕见突变体,在非裔美国人的频率低5倍;③ 几乎所有罕见的CD欧洲变体都是在非裔美国人队列中的欧洲单倍型上发现的,这意味着它们主要由于最近的混血而增加了非裔美国人的疾病风险;④ 欧洲血统的比例与每个非裔美国人携带的罕见CD欧洲变体的数量以及患疾病的多基因风险相关;⑤ 在影响其他10种常见复杂疾病的23种突变中也观察到了类似的发现。
【主编评语】
Genome Medicine近期发表的文章,发现欧洲来源的克罗恩病罕见变种在非裔美国人中更为罕见,其对疾病风险的促进作用主要是由混血造成的,强调了在跨族群遗传评估中考虑血统混合的重要性。(@章台柳)
【原文信息】
The role of admixture in the rare variant contribution to inflammatory bowel disease
2023-11-15, doi: 10.1186/s13073-023-01244-w
国内团队:靶向受体FPR2或可减轻肠炎
EMBO Molecular Medicine——[11.1]
① 筛选并鉴定出一种天然化合物鸽胺(COL)可增强巨噬细胞介导的胞吞作用并减轻肠炎;② COL通过促进LC3相关的吞噬作用(LAP,一种非典型的自噬形式)以增强胞吞作用;③ COL是一种偏向激动剂,可结合参与炎症调节的G蛋白偶联受体甲酰肽受体2(FPR2)的配体结构域;④ Fpr2基因敲除或使用FPR2拮抗剂治疗可消除COL诱导的胞吞作用、抗结肠炎活性和LAP;⑤ FPR2通过调节巨噬细胞中LC3相关的胞吞作用以减轻肠炎,可作为治疗炎症性肠病的潜在靶标。
【主编评语】
胞吞作用(清除凋亡细胞)增强具有消退炎症和修复组织的治疗潜力,然而由于缺乏调节靶标,目前关于增强胞吞作用的药理学方法仍然很少。近日,澳门大学路嘉宏团队联合香港中文大学叶德全团队在EMBO Molecular Medicine上发表文章,发现一种天然化合物鸽胺可通过偏向靶向巨噬细胞上的FPR2来激活LC3相关的胞吞作用并减轻DSS诱导的结肠炎,这项研究通过增强FPR2介导的胞吞作用,为包括结肠炎在内的炎症性疾病提供了一种新的治疗策略。(@圆圈儿)
【原文信息】
Enhancement of efferocytosis through biased FPR2 signaling attenuates intestinal inflammation
2023-11-22, doi: 10.15252/emmm.202317815
Nature:牙釉质与肠道——意想不到的免疫连接
Nature——[64.8]
① 绝大多数患有APS-1患者和乳糜泻患者都会产生抗成釉细胞特异性蛋白的自身抗体(主要是IgA同种型),成釉细胞特异性蛋白的表达是由AIRE在胸腺中诱导的;② 这反过来又导致中枢耐受性的崩溃,随后产生相应自身抗体来干扰牙釉质形成;③ 在乳糜泻中,这种自身抗体的产生似乎是由外周对肠道抗原耐受性的破坏驱动的,而肠道抗原也在釉质组织中表达;④ 这两种情况是先前未确定的IgA依赖性自身免疫性疾病,将其统称为自身免疫性釉质形成不全。
【主编评语】
成釉细胞是颌骨中的特殊上皮细胞,在牙釉质的形成中起着不可或缺的作用——成釉作用。成釉作用依赖于多种成釉细胞衍生的蛋白质,这些蛋白质作为羟基磷灰石晶体的支架。成釉细胞衍生蛋白功能的丧失导致一组罕见的先天性疾病,称为成釉不全。在由AIRE缺陷引起的1型自身免疫性多关节综合征(APS-1)患者和被诊断为乳糜泻的患者中也发现了牙釉质形成缺陷。Nature近期发表的文章,对APS-1和乳糜泻患者如何产生自身免疫性成釉不全的机制进行研究,为相关疾病的诊断和治疗提供了新见解。(@章台柳)
【原文信息】
Autoimmune amelogenesis imperfecta in patients with APS-1 and coeliac disease
2023-11-22, doi: 10.1038/s41586-023-06776-0
Lancet子刊:非乳糜泻性麸质敏感,心理效应不可忽视
Lancet Gastroenterology & Hepatology——[35.7]
① 纳入83名参与者,86%是女性,中位年龄27岁,根据早餐或午餐期望摄入含或不含谷蛋白(E+E–)和实际摄入含或不含谷蛋白(G+G–)食物,随机分为E+G+、E+G–、E–G+和E–G–4组;② E+G+组总体胃肠道症状评分显著高于E-G+和E-G-组,相比早晨,组间整体胃肠道症状的差异在下午更为明显;③ E+G+组腹部不适显著高于E-G+和E-G-组,且下午比上午更显著,E+G+组腹胀显著高于E-G+组;④ E+G+组在精神错乱、头脑模糊和头痛方面显著高于E-G+组。
【主编评语】
许多没有乳糜泻或小麦过敏的人减少了谷蛋白(麸质)的摄入,因为他们认为谷蛋白会导致胃肠道症状。究竟负面预期是否会影响胃肠道和肠道外症状仍然未知。Lancet Gastroenterology & Hepatology上近期刊登的文章,研究了预期谷蛋白摄入与实际谷蛋白摄入对自我报告的非乳糜泻谷蛋白敏感(NCGS)患者胃肠道和肠道外症状的影响。结果表明,预期摄入谷蛋白和实际摄入谷蛋白的组合导致总体胃肠道症状、腹部不适和腹胀最显著,反复接触谷蛋白进一步强化了干预组之间的差异,说明反安慰剂效应在NCGS患者的症状发生中发挥着重要作用。(@RZN)
【原文信息】
The effect of expectancy versus actual gluten intake on gastrointestinal and extra-intestinal symptoms in non-coeliac gluten sensitivity: a randomised, double-blind, placebo-controlled, international, multicentre study
2023-11-28, doi: 10.1016/S2468-1253(23)00317-5
Nature子刊:1型糖尿病模型中肠道菌群和免疫系统相互调节
Nature Communications——[16.6]
① 与野生型NOD小鼠相比,116C-NOD转基因小鼠的1型糖尿病(T1D)发病率显著降低;② 与116C-NOD共养的NOD小鼠T1D发病率低,与肠道菌群变化、肠道通透性降低、T/B细胞亚群变化及Th1向Th17反应转变有关,且肠道菌群特征与T1D相关;③ T细胞改变促进年轻及不同年龄RAG2 KO NOD/116C-NOD小鼠中分节丝状菌增殖;④ 双歧杆菌定植依赖淋巴细胞且在非糖尿病环境中增殖;⑤ 116C-NOD B细胞导致RAG2 KO 116C-NOD小鼠肠道富集Adlercreutzia并降低肠道通透性。
【主编评语】
1型糖尿病(T1D)是一种慢性自身免疫性疾病,且与肠道菌群组成之间存在关联,然而鲜有报道在T1D背景下研究肠道菌群与(先天性和适应性)免疫系统之间的联系。近日,发表在Nature Communications上的这篇文章,利用常用的T1D动物模型NOD小鼠及转基因和敲除变体,对T1D过程中肠道菌群与免疫系统之间的关联进行了实验研究,并发现T1D动物模型中肠道菌群和免疫系统之间存在相互调节。(@圆圈儿)
【原文信息】
Mutual modulation of gut microbiota and the immune system in type 1 diabetes models
2023-11-27, doi: 10.1038/s41467-023-43652-x
感谢本期日报的创作者:章台柳,九卿臣,圆圈儿,RZN,Jadon
点击阅读过去10天的日报:
12-04 | 菌群与癌症治疗,100分综述一文讲透
12-03 | 认知功能/情绪障碍/渐冻症,多文聚焦饮食与脑健康
12-02 | 战肠癌:聚焦筛查和治疗方法新进展
12-01 | 11月,最值得看的30篇肠道健康文献!
11-30 | 《自然·综述》年度回顾:肠神经系统研究的三大进展
11-29 | 菌群寻宝:2篇Nature子刊深挖菌群中的抗菌肽
11-28 | 2篇高分研究,挖掘调节免疫的潜在益生菌
11-27 | 20年大队列新证:吃阿司匹林能否降低癌症风险?
11-26 | Nature子刊:中年开始健康饮食,对寿命的影响有多大?
11-25 | 善事利器:10文一览菌群研究的新方法和新工具
临床研究微生物疗法
2023-06-27
关注并星标CPHI制药在线 6月15日,Abivax递交的「Obefazimod」临床试验申请获CDE受理。Obefazimod(ABX464)是一款口服小分子同类首创药物,其RNA剪接调节作用所产生的效应构成了一种强大的抗炎作用和抗病毒作用,被开发用于治疗多种炎症性疾病(如溃疡性结肠炎、克罗恩病、类风湿性关节炎)和多种病毒性疾病(COVID-19、HIV)。 Obefazimod的抗炎作用是由其结合至靶点帽子结合复合物(CBC)所触发的,CBC位于每个胞内非编码RNA分子的5'端。这种结合可导致一种长的、非编码RNA的剪接,从而诱导单个微小RNA(miRNA)产物miR-124的过度表达。miR-124随后启动一种级联反应,产生强大的抗炎作用。 此外,研究发现Obefazimod与CBC的结合还会增强HIV病毒复制所需的病毒RNA片段的剪接。增强的剪接产生一种新形式的剪接病毒RNA,可能潜在地引发HIV病毒库中HIV感染细胞的免疫识别。 抗病毒方面,临床前数据显示ABX464有潜力减少或消除HIV病毒的病毒储备,诱导长期病毒载量的控制,防止耐药性HIV突变体的出现。已公布的2a期临床研究结果显示:在完全抑制的HIV患者中,ABX464治疗可降低外周血CD4+ T细胞中HIV DNA总量,同时还会降低直肠组织CD45+T细胞中HIV DNA总量。 抗炎方面,Obefazimod针对溃疡性结肠炎(UC)的临床试验已进入3期阶段,针对活动性类风湿关节炎(RA)和克罗恩病(CD)的临床试验已进入2期阶段。 Obefazimod有望成为UC患者新选择 治疗UC方面,Obefazimod表现不错。2018年9月公布的Obefazimod用于中重度UC患者诱导治疗的2a期临床试验结果显示:针对免疫调节剂失败的中重度UC患者,接受Obefazimod治疗8周后,35%的患者获得了临床缓解。在粘膜愈合方面,50%的患者出现了积极反应。而且,在治疗4周后,与安慰剂相比,接受Obefazimod治疗患者的钙结合蛋白显著降低(钙结合蛋白是UC中研究最完备的生物标志物)。 2020年9月公布的Obefazimod用于UC维持治疗的2a期临床研究数据显示:针对先前对至少一种现有疗法不耐受和/或难治的UC患者,经过一年Obefazimod维持治疗,69%的患者获得临床缓解,94%的患者受益于临床应答,44%患者处于内镜缓解。 2022年9月在The Lancet Gastroenterology & Hepatology杂志上发布的Obefazimod用于UC诱导治疗的2b期临床试验结果显示:与安慰剂相比,所有Obefazimod组(25 毫克、50 毫克和 100 毫克组)均显著改善了患有中重度活动性UC患者的状况。而且,Obefazimod组也达到了所有关键的次要终点,包括内窥镜改善、临床缓解、临床反应和粪便钙卫蛋白减少等。 2023年4月公布的Obefazimod针对UC的2b期维持研究的两年疗效和安全性数据显示:217名UC患者经过每天一次口服50mg的Obefazimod治疗两年(96周)后,52.5%(n=114)的患者出现临床缓解,59.0%(n=128)和35.9%(n=78)的患者分别实现了内窥镜改善和内窥镜缓解;在8周诱导研究后未显示临床缓解的患者亚组中,48.2%(n=81)的患者在治疗两年后显示了新的临床缓解;安全性和耐受性状况与之前的结果一致,没有观察到相关的安全问题。 上述试验数据表明,Obefazimod有望成为中重度UC患者诱导治疗和维持治疗的新选择。而且,Obefazimod使用方便,仅需每日口服一次。 UC新药不断涌现 溃疡性结肠炎(UC)是一种慢性、非特异性肠道炎症性疾病,以结肠黏膜连续性、弥漫性炎症改变为特点。UC常见的临床表现为持续或反复发作的腹泻、黏液脓血便伴腹痛、里急后重和不同程度的全身症状,病程多在 4~6 周以上,其中黏液脓血便是UC最常见的症状。按严重程度,UC可分为轻、中、重度三级,不同级别治疗方案也不同。 据统计,UC影响全世界数百万人,严重影响患者的生活品质,亟需新疗法。传统UC治疗药物柳氮磺吡啶 (SASP)、 5-氨基水杨酸(5-ASA)制剂(巴柳氮、美沙拉秦和奥沙拉秦)、激素(可的松、泼尼松龙、地塞米松等)、免疫抑制剂(如巯唑嘌呤、环孢素、他克莫司等)和生物制剂(如阿达木单抗、英夫利昔单抗)等虽然对UC有效,但长期使用疗效有限,或者副作用大。 近年来,全球监管机构批准了多款UC治疗新药,详见下表。除了TNF抑制剂,新获批的UC新药涉及单抗和小分子化药,相关药物靶点包括JAK、S1PR、IL-23、α4整合素等。 此外,目前全球还有多款在研UC新药,详见下表。其中S1PR调节剂Etrasimod治疗中重度活动性UC的新药申请已于2022年12月获FDA受理,预计今年下半年将获批。我国恒瑞医药JAK1抑制剂艾玛昔替尼进展也较快,其UC适应症已进入3期临床。期待为了有更多的UC新药获批上市,造福广大患者。【企业推荐】智药研习社近期课程报名来源:CPHI制药在线声明:本文仅代表作者观点,并不代表制药在线立场。本网站内容仅出于传递更多信息之目的。如需转载,请务必注明文章来源和作者。投稿邮箱:Kelly.Xiao@imsinoexpo.com▼更多制药资讯,请关注CPHI制药在线▼点击阅读原文,进入智药研习社~
临床2期临床3期临床结果
100 项与 奥沙拉秦钠 相关的药物交易
登录后查看更多信息
研发状态
10 条最早获批的记录, 后查看更多信息
登录
| 适应症 | 国家/地区 | 公司 | 日期 |
|---|---|---|---|
| 溃疡性结肠炎 | 美国 | 1990-07-31 |
登录后查看更多信息
临床结果
临床结果
适应症
分期
评价
查看全部结果
| 研究 | 分期 | 人群特征 | 评价人数 | 分组 | 结果 | 评价 | 发布日期 |
|---|
No Data | |||||||
登录后查看更多信息
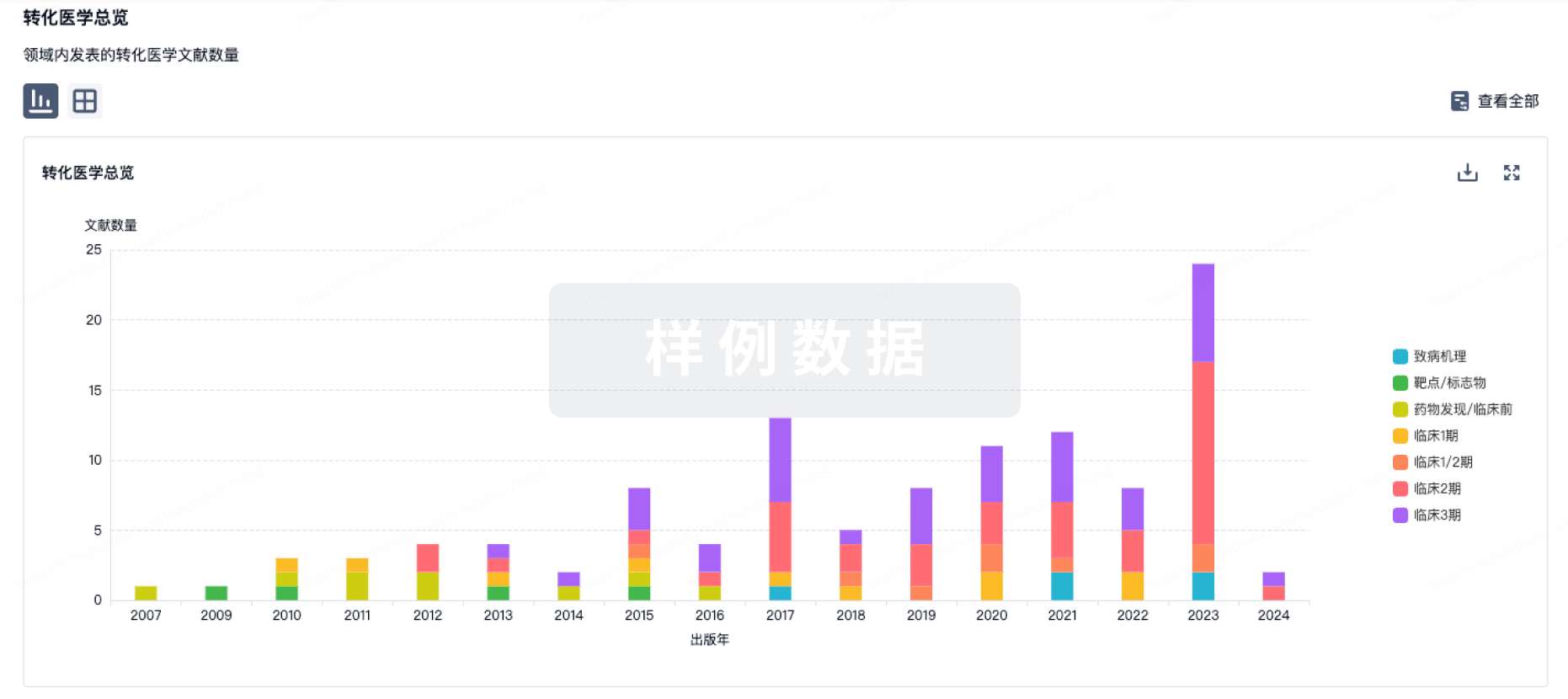
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
核心专利
使用我们的核心专利数据促进您的研究。
登录
或
临床分析
紧跟全球注册中心的最新临床试验。
登录
或
批准
利用最新的监管批准信息加速您的研究。
登录
或
特殊审评
只需点击几下即可了解关键药物信息。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用




