预约演示
更新于:2025-05-07
HCAR3
更新于:2025-05-07
基本信息
别名 G protein-coupled receptor 109B、G-protein coupled receptor 109B、G-protein coupled receptor HM74 + [10] |
简介 Receptor for 3-OH-octanoid acid mediates a negative feedback regulation of adipocyte lipolysis to counteract prolipolytic influences under conditions of physiological or pathological increases in beta-oxidation rates. Acts as a low affinity receptor for nicotinic acid. This pharmacological effect requires nicotinic acid doses that are much higher than those provided by a normal diet. |
关联
10
项与 HCAR3 相关的药物作用机制 HCAR3激动剂 [+1] |
最高研发阶段批准上市 |
首次获批国家/地区 日本 |
首次获批日期1956-11-01 |
作用机制 HCAR3激动剂 [+1] |
在研机构- |
原研机构- |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
作用机制 HCAR3激动剂 [+2] |
在研机构 |
原研机构 |
非在研适应症- |
最高研发阶段临床3期 |
首次获批国家/地区 美国 |
首次获批日期2008-02-15 |
212
项与 HCAR3 相关的临床试验NCT06843148
Stimulating Adipose Tissue Fatty Acid Disposal with Low-dose, Postprandial, Intermittent Niacin for the Treatment of Metabolic Dysfunction-associated Steatotic Liver Disease (MASLD).
Metabolic dysfunction-associated steatotic liver disease (MASLD) (aka non-alcoholic fatty liver disease), commonly occurring in individuals with obesity and type 2 diabetes can lead to liver inflammation/ fibrosis. MASLD results from fat being disproportionately deposited in the liver.
The goal of this mechanistic study is to investigate metabolic response in patients aged 50 to 80 years with non-alcoholic fatty liver disease, after niacin (vitamin B3) treatment.
The main questions it aims to answer are:
* Does Niacin lower the fat deposition in the liver?
* Does Niacin raise White Adipose Tissue storage of dietary fatty acids?
Researchers will compare Niacin to a placebo (a look-alike substance that contains no drug) to compare the metabolic response.
Duration of study per participant: Up to 28 weeks
The goal of this mechanistic study is to investigate metabolic response in patients aged 50 to 80 years with non-alcoholic fatty liver disease, after niacin (vitamin B3) treatment.
The main questions it aims to answer are:
* Does Niacin lower the fat deposition in the liver?
* Does Niacin raise White Adipose Tissue storage of dietary fatty acids?
Researchers will compare Niacin to a placebo (a look-alike substance that contains no drug) to compare the metabolic response.
Duration of study per participant: Up to 28 weeks
开始日期2025-04-01 |
申办/合作机构 |
NCT06858332
Lipoprotein(a) Levels in Patients With Atherosclerotic Cardiovascular Diseases in Russia
This study has the purpose to answer how the Lipoprotein(a) (Lp(a)) level is distributed among Atherosclerotic cardiovascular disease (ASCVD) patients in Russia, and what is the connection between elevated levels of this parameter and the cardiovascular disease (CVD) risk.
开始日期2025-03-31 |
NCT06582706
Nicotinic Acid for the Treatment of Alzheimer's Disease
Increased dietary intake of niacin is correlated with reduced risk of Alzheimer's Disease and age-associated cognitive decline. The goal of this study is to collect data on the penetration of commercially available, FDA approved, extended-release niacin into the spinal fluid. One dose of 500 mg nicotinic acid will be used (in addition to placebo) to build a dose response curve for this compound in human cerebrospinal fluid. This objective will demonstrate target engagement of HCAR2 in the central nervous system, after oral treatment with niacin. The primary endpoints are to show increased nicotinic acid levels in blood and cerebrospinal fluid. A secondary endpoint is to collect safety and tolerability data of niacin in this particular population.
开始日期2024-12-19 |
申办/合作机构  Indiana University Indiana University [+1] |
100 项与 HCAR3 相关的临床结果
登录后查看更多信息
100 项与 HCAR3 相关的转化医学
登录后查看更多信息
0 项与 HCAR3 相关的专利(医药)
登录后查看更多信息
86
项与 HCAR3 相关的文献(医药)2025-06-01·Biochimica et Biophysica Acta (BBA) - Molecular Cell Research
Activation and signaling characteristics of the hydroxy-carboxylic acid 3 receptor identified in human neutrophils through a microfluidic flow cell technique
Article
作者: Li, Wenyan ; Dahlgren, Claes ; Levin, Neele K ; Karlsson, Anders ; Sundqvist, Martina ; Karlsson, Roger ; Forsman, Huamei
2024-11-01·Cell Reports
Structural basis for ligand recognition of the human hydroxycarboxylic acid receptor HCAR3
Article
作者: Qian, Chungen ; Pan, Xin ; Zhu, Lizhe ; Li, Xinyu ; Liu, Aijun ; Zhang, Binghao ; Chen, Geng ; Ning, Peiruo ; Zhang, Zhiyi ; Wang, Qinggong ; Gong, Kaizheng ; Xiang, Xufu ; Du, Yang ; Li, Jiancheng ; Ye, Fang
2024-10-01·Anticancer Research
Identification of New Potential Targets for Pancreatic Ductal Adenocarcinoma by Integrated Bioinformatic Analysis
Article
作者: Ren, K E ; Li, Rujia ; Zeng, Yuhao ; LI, JUN ; LI, RUJIA ; ZENG, YUHAO ; Yang, Ting ; Li, Jun ; REN, KE ; NAGAKAWA, YUICHI ; Yi, Shuang-Qin ; YI, SHUANG-QIN ; NATSUYAMA, YUTARO ; YANG, TING ; Nagakawa, Yuichi ; Natsuyama, Yutaro
1
项与 HCAR3 相关的新闻(医药)2023-09-02
最新资讯8月28日,信达生物制药集团 (信达生物) 官网发布公告称,抗PCSK9单克隆抗体信必乐®(托莱西单抗注射液)在获得国家药品监督管理局(NMPA)批准后,8月25日开始正式正式向全国各大医院和药房供药,并迅速在北京大学第一医院,复旦大学附属中山医院等医院开出了全国首批处方。 “PCSK9抑制剂作为一种新型降胆固醇药物,能强效降低LDL-C水平且安全性良好。托莱西单抗在多个注册III期研究中展现出强大的降脂疗效和良好的安全性,与国内已上市PCSK9单抗相比,具有更长的给药间隔。尽管目前临床上已有多种降脂药物,但中国患者血脂整体达标情况不佳,相信托莱西单抗的上市会提升PCSK9单抗在降脂治疗方面的可及性,为广大高胆固醇血症及混合型血脂异常患者提供优质的治疗选择。” 北京大学第一医院霍勇教授表示: 引此前8月中旬消息,信达生物的托莱西单抗注射液获得国家药品监督管理局(NMPA)批准,用于治疗原发性高胆固醇血症(包括杂合子型家族性和非家族性高胆固醇血症)和混合型血脂异常的成人患者。作为中国首个获批的本土自主研发PCSK9抑制剂,托莱西单抗注射液是信达生物布局心血管疾病领域的首款产品,也是信达生物成立12年来的第十款产品。 “与其他已获批的PCSK9单抗相比,托莱西单抗有更长给药间隔的优势,同时观察到脂蛋白a的明显下降。作为国内首个具有本土自主知识产权的PCSK9单抗,托莱西单抗的临床开发凝聚了国内多位心血管领域专家的心血,也证明了信达生物在心血管药物开发和临床开发领域的实力,以及监管机构对其疗效和安全性的认可。托莱西单抗的获批标志着信达生物在心血管及代谢(CVM)领域的前瞻布局开始进入收获期,信达生物将秉持着‘开发出老百姓用得起的高质量生物药’的使命,将为更广阔的慢病患者提供更多更优的治疗选择。” 信达生物制药集团临床开发副总裁钱镭博士表示: 高胆固醇血症高胆固醇血症(hypercholesterolemia),是指血液中胆固醇水平异常升高的一种状况。胆固醇是一种脂质类物质,它在人体中扮演着重要的角色,包括细胞膜的构建、激素合成和胆汁酸的产生等。当血液中的胆固醇水平持续升高时,可能会增加心血管疾病(如冠心病、中风及动脉粥样硬化等)的风险。由于胆固醇不溶于水,它在血浆中通过脂蛋白颗粒运输。脂蛋白按密度分为:极低密度脂蛋白(VLDL)、低密度脂蛋白(LDL)、中密度脂蛋白(IDL)、和高密度脂蛋白(HDL)。所有脂蛋白都携带胆固醇,较高水平的高密度脂蛋白胆固醇具有保护作用,而低密度脂蛋白胆固醇水平升高与动脉粥样硬化和冠心病风险增加有关。根据此前文章推介,重磅解读:IQVIA 发布全球肥胖浪潮下心血管代谢创新药的复兴,IQVIA 发布的代谢性疾病相关市场调研,高水平低密度脂蛋白胆固醇 (LDL-C) 已经成为全球范围内死亡率和发病率的主要风险因素之一。在正常情况下,位于肝细胞表面的低密度脂蛋白受体(LDLR) 与 APOB/LDL-C 结合形成复合物,并通过 LDLR 与 LDLRAP1 的相互作用,复合物被内吞进入细胞。当复合物在细胞内解离时,LDLR 通常会回收到细胞质膜,游离胆固醇则会在细胞内消耗掉。PCSK9 通过细胞表面的相互作用介导 LDLR 的活性抑制。来源于,DOI: 10.3389/fgene.2020.572045在遗传性高胆固醇血症中,LDLR、APOB、PCSK9 和 LDLRAP1 的单个突变可导致从轻微到严重的多种表型。LDLR 突变会影响 LDL-C 与 LDLR 的结合,导致 LDL-C 在血液循环中蓄积。APOB 基因突变会导致蛋白质功能障碍,使 LDL-C 与 LDLR 的结合受到限制。PCSK9 基因突变可导致:(1) PCSK9 活性增加;(2) 没有 LDLR 循环;(3) 血浆中 LDL-C 水平累积。在这种情况下,抗 PCSK9 疗法,包括新出现的 PCSK9 抑制剂(如 alirocumab、evolocumab),可用于降低 LDL-C,预防心血管疾病。高胆固醇血症相关药研现状根据 智慧芽新药情报中心 统计,高胆固醇血症及其相关的代谢性疾病是全球药企的热门研究适应症,共有337个高胆固醇血症药物,来自311个机构,覆盖71个靶点,开展1872个临床试验。在适应症高胆固醇血症下,HMG-CoA reductase 是最常见的临床靶点,有7个药物已经获得批准上市,还有20个药物处于非在研阶段。其次是PCSK9靶点,有4个药物获得批准上市,13个药物处于临床前阶段,21个药物处于非在研阶段。以靶向 PCSK9 的药物为例,包括开篇提到的信达生物的托莱西单抗 (Tafolecimab)、Novartis的英克司兰钠 (Inclisiran)、Sanofi 和 Regeneron 的阿利西尤单抗 (Alirocumab) 以及安进公司的依洛尤单抗 (Evolocumab) 均已获批上市用于高胆固醇血症或血脂障碍等相关疾病。另有多款药物处于申请上市阶段。在GPCR 类靶点方面,热门靶点包括NIACR1、AT1R、HCAR3、LTRs、PGD2R、GPR120等,成为治疗高胆固醇血症的临床治疗靶点。其中,NIACR1 激动剂 Acipimox、H1/H3 受体双重拮抗剂 Betahistine、HCAR3/NIACR1 双重激动剂 Niacin 均已获批上市用于治疗高胆固醇血症、动脉粥样硬化、脂质代谢紊乱等相关疾病。诺华公司的小干扰核酸药物: Inclisiran8月22日,诺华公司 (Novartis)的英克司兰钠宣布其长效降脂小干扰核酸(siRNA)降胆固醇药物英克司兰钠(Inclisiran)注射液获得中国国家药监局批准,用于成人原发性高胆固醇血症或混合性血脂异常患者的治疗。据悉,Inclisiran是目前全球首款也是唯一一款用于降低低密度脂蛋白胆固醇(LDL-C)的siRNA疗法,患者每年进行两次注射即可降低LDL-C。由于这款药物的长效性,患者无需每天服药,因此inclisiran也被称为心血管领域的“疫苗”。8月28日,诺华官网再次重磅公布 ORION-8 的最新长期数据,ORION-8 是 ORION-9、ORION-10、ORION-11 和 ORION-3 试验的 III 期开放标签扩展试验。这些数据表明,Leqvio (Inclisiran) 在他汀类药物治疗的基础上,通过每年两次给药,可在动脉粥样硬化性心血管疾病 (ASCVD) 或杂合性家族性高胆固醇血症 (HeFH)1 患者中持续降低低密度脂蛋白胆固醇 (LDL-C),并可持续六年以上。Merck 公司的口服PCSK9 抑制剂: MK-06168月25日,默克公司 (Merck) 官网发布公告称,正式启动公司的一项3期临床项目CORALreef。该项目旨在研究MK-0616,一款正在研究的口服 PCSK9 抑制剂,用于治疗成人高胆固醇血症。据悉,这是首个口服 PCSK9 抑制剂的 3 期临床项目。目前,首批参与者正在参加两项评估 MK-0616 降低低密度脂蛋白(LDL-C) 的注册 3 期研究:CORALreef Lipids 和 CORALreef HeFH。默克还计划在2023年底前启动一项3期心血管结果研究--CORALreef Outcomes。下图为 www.clinicaltrials.gov 上关于 MK-0616 的临床研究情况: 以上文章来源于GPCR drug diacovery,作者段思研 本文仅作信息交流之目的,文中观点不代表相关疾病治疗立场,也不是治疗方案推荐。如需获得治疗方案指导,请前往正规医院就诊。
上市批准申请上市临床3期
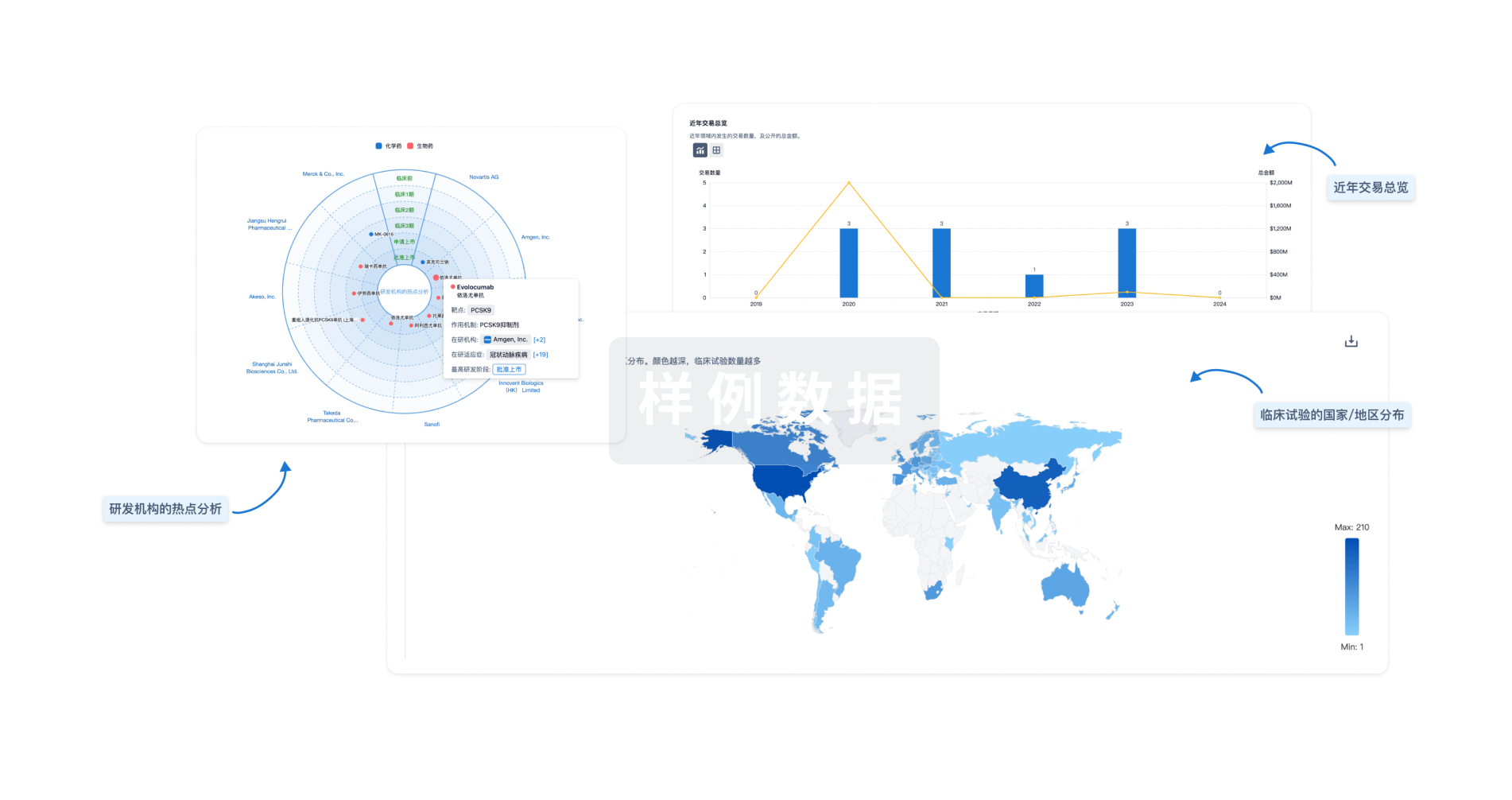
分析
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用




