预约演示
更新于:2025-03-08

BenevolentAI Ltd.
更新于:2025-03-08
概览
标签
神经系统疾病
其他疾病
内分泌与代谢疾病
小分子化药
疾病领域得分
一眼洞穿机构专注的疾病领域
暂无数据
技术平台
公司药物应用最多的技术
暂无数据
靶点
公司最常开发的靶点
暂无数据
| 排名前五的药物类型 | 数量 |
|---|---|
| 小分子化药 | 6 |
关联
6
项与 BenevolentAI Ltd. 相关的药物作用机制 NTRK抑制剂 [+3] |
非在研适应症- |
最高研发阶段临床1/2期 |
首次获批国家/地区- |
首次获批日期- |
靶点 |
作用机制 PDE10A抑制剂 |
非在研适应症- |
最高研发阶段临床1期 |
首次获批国家/地区- |
首次获批日期- |
作用机制 RARα激动剂 [+1] |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
4
项与 BenevolentAI Ltd. 相关的临床试验NCT06118385
A Randomised, Double-blind, Placebo-controlled, Phase 1 First-in-human Study to Investigate the Safety, Tolerability, Pharmacokinetics, and Food Effect of Single- and Multiple-ascending Doses of BEN8744 in Healthy Subjects.
BEN8744 is an experimental new medicine for treating inflammatory bowel diseases such as Ulcerative Colitis.
The study will test single and repeated doses of BEN8744 or placebo by mouth. BEN8744 is a first in human study, so will start with a small dose and the dose will be increased as the study progresses. The goal is to find out its side effects and blood levels when taken by mouth and whether food affects the blood levels.
This is a 3-part study (Parts A, B and C) in up to 108 healthy people, aged 18-65.
Part A, will include up to 64 participants, single doses of BEN8744 or placebo. They'll take about 2 weeks to finish the study, stay on the ward for 4 nights and 5 days in a row and make 2 outpatient visits.
Part B, will include up to 12 participants, single doses of BEN8744 with and without food. They'll take up to 3 weeks to finish the study, stay on the ward for 4 nights and 5 days in a row on 2 occasions, and make 2 outpatient visits.
Part C will include up to 32 participants repeat doses of the BEN8744 or placebo for 14 days. They'll take about 4 weeks to complete the study, stay on the ward for 17 nights and 18 days in a row and make 2 outpatient visits.
The study will test single and repeated doses of BEN8744 or placebo by mouth. BEN8744 is a first in human study, so will start with a small dose and the dose will be increased as the study progresses. The goal is to find out its side effects and blood levels when taken by mouth and whether food affects the blood levels.
This is a 3-part study (Parts A, B and C) in up to 108 healthy people, aged 18-65.
Part A, will include up to 64 participants, single doses of BEN8744 or placebo. They'll take about 2 weeks to finish the study, stay on the ward for 4 nights and 5 days in a row and make 2 outpatient visits.
Part B, will include up to 12 participants, single doses of BEN8744 with and without food. They'll take up to 3 weeks to finish the study, stay on the ward for 4 nights and 5 days in a row on 2 occasions, and make 2 outpatient visits.
Part C will include up to 32 participants repeat doses of the BEN8744 or placebo for 14 days. They'll take about 4 weeks to complete the study, stay on the ward for 17 nights and 18 days in a row and make 2 outpatient visits.
开始日期2023-08-30 |
申办/合作机构 |
NCT04737304
A First-in-Human, Double-Blind, Randomised, Vehicle Controlled Phase I/II Proof of Concept Study to Investigate the Safety, Tolerability, Pharmacokinetics and Efficacy of BEN2293 in Patients With Mild to Moderate Atopic Dermatitis
A randomised, adaptive design, double-blind, placebo-controlled, first-in-human, two-part study to investigate the safety, tolerability, PK and preliminary efficacy of multiple topical doses of BEN2293 in patients with mild to moderate AD.
开始日期2020-10-14 |
申办/合作机构 |
NCT03194217
Dose Finding Phase IIb Study of Bavisant to Evaluate Its Safety and effiCacy in treAtment of exceSsive Daytime sleePiness (EDS) in PARkinson's Disease (PD).
This phase 2b study is designed as multicentre, multinational, randomized, double blind, parallel group and placebo controlled with three doses of Bavisant (0.5, 1, and 3 mg/d) in subjects with excessive daytime sleepiness with Parkinson's disease.
开始日期2017-11-10 |
申办/合作机构 |
100 项与 BenevolentAI Ltd. 相关的临床结果
登录后查看更多信息
0 项与 BenevolentAI Ltd. 相关的专利(医药)
登录后查看更多信息
3
项与 BenevolentAI Ltd. 相关的文献(医药)2024-12-20·ORGANIC PROCESS RESEARCH & DEVELOPMENT
Process Development for the Manufacture of a Topical Pan-Trk Inhibitor Incorporating Decarboxylative sp2–sp3 Cross-Coupling
作者: Moore, Jonathan ; McGuffog, Jane ; Fengas, David ; Ashwood, Michael S. ; Robas, Nicola M. ; Stevenson, Neil G. ; Balmond, Edward I. ; Wise, Lisa
The development of a synthetic route toward topical pan-Trk inhibitor 1 is described as an eight-stage synthesis from available starting materials.Process improvements include the development of a decarboxylative sp2-sp3 cross-coupling which had not previously been demonstrated on scale.Parameters were explored, balancing the safety aspects with conversion and selectivity, scaling up in a stepwise fashion to multiple successful 0.7 kg batches.The cross-coupling showed high diastereoselectivity, with the opposite diastereomer not observed in the crude 19F NMR.Selectivity was further improved by crystallizing the downstream pyrrolidine salt after Boc deprotection, to give a diastereomer ratio of 99.5:0.5 by UPLC.This route has been reproducibly demonstrated in two GMP campaigns delivering API on kilogram scale, in >98% area purity by HPLC.The route design, solid-form screening, process research, and manufacture have enabled crucial first-in-human (FIH) clin. studies, through focus on speed of delivery.
2022-03-24·The Journal of clinical endocrinology and metabolism2区 · 医学
Identification of Rare Loss-of-Function Genetic Variation Regulating Body Fat Distribution
2区 · 医学
Article
作者: Langenberg, Claudia ; Bowker, Nicholas ; Saudek, Vladimir ; Small, Kerrin S ; O’Rahilly, Stephen ; Stewart, Isobel D ; Semple, Robert K ; Griffin, John D ; Luan, Jian’an ; Dong, Liang ; Perry, John R B ; Barroso, Inês ; Rocha, Nuno ; Glastonbury, Craig A ; Savage, David B ; Scott, Robert A ; McCarthy, Mark I ; Wheeler, Eleanor ; Lotta, Luca A ; Van de Streek, Marcel ; Li, Chen ; Wittemans, Laura B L ; Cai, Lina ; Patel, Satish ; Lucarelli, Debora M E ; Day, Felix R ; Kerrison, Nicola D ; Zhao, Yajie ; Wareham, Nicholas J ; Koprulu, Mine
Abstract:
Context:
Biological and translational insights from large-scale, array-based genetic studies of fat distribution, a key determinant of metabolic health, have been limited by the difficulty in linking predominantly noncoding variants to specific gene targets. Rare coding variant analyses provide greater confidence that a specific gene is involved, but do not necessarily indicate whether gain or loss of function (LoF) would be of most therapeutic benefit.
Objective:
This work aimed to identify genes/proteins involved in determining fat distribution.
Methods:
We combined the power of genome-wide analysis of array-based rare, nonsynonymous variants in 450 562 individuals in the UK Biobank with exome-sequence-based rare LoF gene burden testing in 184 246 individuals.
Results:
The data indicate that the LoF of 4 genes (PLIN1 [LoF variants, P = 5.86 × 10–7], INSR [LoF variants, P = 6.21 × 10–7], ACVR1C [LoF + moderate impact variants, P = 1.68 × 10–7; moderate impact variants, P = 4.57 × 10–7], and PDE3B [LoF variants, P = 1.41 × 10–6]) is associated with a beneficial effect on body mass index–adjusted waist-to-hip ratio and increased gluteofemoral fat mass, whereas LoF of PLIN4 (LoF variants, P = 5.86 × 10–7 adversely affects these parameters. Phenotypic follow-up suggests that LoF of PLIN1, PDE3B, and ACVR1C favorably affects metabolic phenotypes (eg, triglycerides [TGs] and high-density lipoprotein [HDL] cholesterol concentrations) and reduces the risk of cardiovascular disease, whereas PLIN4 LoF has adverse health consequences. INSR LoF is associated with lower TG and HDL levels but may increase the risk of type 2 diabetes.
Conclusion:
This study robustly implicates these genes in the regulation of fat distribution, providing new and in some cases somewhat counterintuitive insight into the potential consequences of targeting these molecules therapeutically.
BMJ
Clinical trial design and dissemination: comprehensive analysis of clinicaltrials.gov and PubMed data since 2005
Article
作者: Zwierzyna, Magdalena ; Hingorani, Aroon D ; Davies, Mark ; Hunter, Jackie
OBJECTIVE:
To investigate the distribution, design characteristics, and dissemination of clinical trials by funding organisation and medical specialty.
DESIGN:
Cross sectional descriptive analysis.
DATA SOURCES:
Trial protocol information from clinicaltrials.gov, metadata of journal articles in which trial results were published (PubMed), and quality metrics of associated journals from SCImago Journal and Country Rank database.
SELECTION CRITERIA:
All 45 620 clinical trials evaluating small molecule therapeutics, biological drugs, adjuvants, and vaccines, completed after January 2006 and before July 2015, including randomised controlled trials and non-randomised studies across all clinical phases.
RESULTS:
Industry was more likely than non-profit funders to fund large international randomised controlled trials, although methodological differences have been decreasing with time. Among 27 835 completed efficacy trials (phase II-IV), 15 084 (54.2%) had disclosed their findings publicly. Industry was more likely than non-profit trial funders to disseminate trial results (59.3% (10 444/17 627) v 45.3% (4555/10 066)), and large drug companies had higher disclosure rates than small ones (66.7% (7681/11 508) v 45.2% (2763/6119)). Trials funded by the National Institutes of Health (NIH) were disseminated more often than those of other non-profit institutions (60.0% (1451/2417) v 40.6% (3104/7649)). Results of studies funded by large drug companies and NIH were more likely to appear on clinicaltrials.gov than were those from non-profit funders, which were published mainly as journal articles. Trials reporting the use of randomisation were more likely than non-randomised studies to be published in a journal article (6895/19 711 (34.9%) v 1408/7748 (18.2%)), and journal publication rates varied across disease areas, ranging from 42% for autoimmune diseases to 20% for oncology.
CONCLUSIONS:
Trial design and dissemination of results vary substantially depending on the type and size of funding institution as well as the disease area under study.
62
项与 BenevolentAI Ltd. 相关的新闻(医药)2024-12-11
Plus, news about NewAmsterdam, Rocket, ImmunityBio, CG Oncology, Spruce Biosciences, Pharming, Veradermics, Noema Pharma, Ambrosia Biosciences, EsoBiotec, Cellectar Biosciences and Hepion:
BenevolentAI’s overhaul:
The struggling AI biotech
said
it plans to lay off staff, consider delisting from the Euronext Amsterdam stock exchange, partner its early-stage assets and undergo other additional organizational shifts as
co-founder Ken Mulvany returns
. Its Amsterdam-listed stock was down 10% on Wednesday.
— Kyle LaHucik
Cimeio Therapeutics, Kyowa Kirin make a deal:
The Japanese drugmaker
is paying
up to $300 million as part of an agreement to develop new cell therapies. Additional financial terms were not disclosed.
— Jaimy Lee
At least three public offerings have been disclosed over the past day, including
NewAmsterdam
’s
bid
to raise $300 million;
Rocket
’s upsized $165 million
offering
;
ImmunityBio
’s $100 million
financing
; and
CG Oncology
’s
offering
, which has yet to be priced.
— Jaimy Lee
Spruce’s Phase 2b fail for endocrine disorder drug:
The South San Francisco biotech said tildacerfont administered once daily did not
meet
the primary endpoint of glucocorticoid reduction in a mid-stage study of people with adult congenital adrenal hyperplasia. But the trial’s principal investigator said the data suggest twice daily dosing may be more effective. Its stock
$SPRB
was down about 25% on Wednesday morning.
— Ayisha Sharma
Pharming’s rare disease med passes Phase 3 test in kids:
The Dutch drugmaker’s Joenja for activated phosphoinositide 3-kinase delta syndrome (APDS)
met
the primary endpoints in a late-stage trial of patients aged four to 11 years old. Joenja
won
FDA approval for APDS patients aged 12 and older in March 2023. Pharming plans to submit global regulatory filings in children starting in 2025.
— Ayisha Sharma
Veradermics’ $75M Series B:
The Connecticut-based dermatology and aesthetics biotech raised the
financing
as it begins a Phase 2/3 study in hair loss.
— Kyle LaHucik
Noema adds $35M from EQT Life Sciences:
A second close of a Series B
first announced in March of last year
brings the total for the round to CHF 130 million, which is about $147 million. The cash will fund Noema’s four ongoing Phase 2 trials: mGluR5 negative allosteric modulator basimglurant in trigeminal neuralgia and seizures in tuberous sclerosis complex; gemlapodect, a PDE10a inhibitor, in Tourette syndrome; and NOE-115, a monoamine modulator, in menopause symptoms.
— Elizabeth Cairns
Merck backs Ambrosia:
The New Jersey pharma giant invested in the biotech’s Series A, which now
totals
$25 million.
Ambrosia
previously announced plans to take over a former Pfizer site in Boulder as it builds out an oral small-molecule pipeline for obesity and metabolic disorders.
— Kyle LaHucik
EsoBiotec scores €22M:
The Belgian cell therapy specialist
raised
the equivalent of $23 million from backers that include Thuja Capital, UCB Ventures and Invivo Partners. The company’s immune-shielded lentiviral vector, which is called ESO-T01, is designed to reprogram T lymphocytes into BCMA CAR-T cells
in vivo
. It will enter an investigator-initiated trial in multiple myeloma in China, its first in humans, and data are expected in the second half of 2025, which could allow expansion to indications including autoimmune diseases.
— Elizabeth Cairns
Cellectar lays off 60% of its workers:
The biotech is
prioritizing
its pipeline and letting go of staff. Cellectar had 20 employees at the end of 2023. Its stock price
$CLRB
sank 62% in pre-market trading on Wednesday.
— Kyle LaHucik
Hepion Pharmaceuticals
ends
merger plans with Pharma Two B.
— Jaimy Lee
细胞疗法临床3期临床2期上市批准临床结果
2024-12-11
Artificial intelligence biotech firm BenevolentAI is reorganizing its operations for the second time in less than two years, and this time it says the changes will take the company back to its original mission of serving biopharma industry partners.
Going forward, London-based BenevolentAI will refocus its efforts on its core technologies for drug discovery and development. The Wednesday announcement offered few financial details, but the company did say the coming changes include plans to take the company private.
BenevolentAI went public in a 2022 SPAC merger that brought the company to the Euronext Amsterdam exchange and infused it with €225 million (about $236 million). At that time, the company said it had 300 employees spread across offices in London and New York as well as laboratories in Cambridge, U.K. BenevolentAI also said its internal pipeline had more than 20 programs in various stages of development. Since its public markets debut, the company’s stock price has steadily declined.
Sponsored Post
The Funding Model for Cancer Innovation is Broken — We Can Fix It
Closing cancer health equity gaps require medical breakthroughs made possible by new funding approaches.
By Eve McDavid - CEO of Mission-Driven Tech
BenevolentAI had taken a two-pronged approach to business. It sought partnerships with pharmaceutical and biotech firms interested in using its proprietary technologies for identifying or validating drug targets, designing new drugs, and repurposing existing drugs for other indications. Companies leveraging BenevolentAI’s technologies for such applications include Merck KGaA, AstraZeneca, and Eli Lilly. BenevolentAI receives upfront fees and milestone payments. In the case of Lilly, the AI company received an equity investment.
The second piece of BenevolentAI’s business model focused on in-house drug discovery and development with a goal of out licensing or partnering these assets at key value-inflection points. This part of the business has had some stumbles. Last year, BenevolentAI stopped work on an internally discovered atopic dermatitis program after Phase 2 results showed the drug did not reduce itch and inflammation.
A subsequent corporate restructuring focused the company on other clinical and preclinical assets. Months later, CEO Joanna Shields stepped down from her role. She was succeeded by Bayer veteran Joerg Moeller. In October, BenevolentAI announced the departure of Moeller and the appointment of Kenneth Mulvaney as executive chairman. Mulvaney founded BenevolentAI in 2013 and he returned to the company’s board of directors this past May.
Currently, BenevolentAI’s most advanced internal program is BEN-8744, an orally administered PDE10 inhibitor. This drug is early clinical development for ulcerative colitis and the company says the molecule has potential applications in other inflammatory bowel disorders. Preclinical assets include drug candidates for amyotrophic lateral sclerosis and glioblastoma. While BenevolentAi said it will maintain its drug development partnering strategy for internally discovered and developed drugs, it will now seek to partner these assets earlier in the development cycle. On the technology front, BenevolentAI said it will transform its offerings into more flexible, standalone products designed to address the needs of its biopharma partners.
Consumer / Employer
Health Executives on Digital Transformation in Healthcare
Hear executives from Quantum Health, Surescripts, EY, Clinical Architecture and Personify Health share their views on digital transformation in healthcare.
By MedCity News
BenevolentAI did not specify how many jobs could be cut in the reorganization. The company’s financial report for the first half of 2024 states it had 180 employees in London and Cambridge. As of June 30, BenevolentAI reported its cash position was £38.1 million (about $48.5 million), which the company had projected would support operations late into the third quarter of 2025. With the restructuring, BenevolentAI now projects its cash runway will extend into 2027. BenevolentAI said its board concluded it is in the company’s best interest to delist from Euronext Amsterdam because the financial costs associated with being a public company divert resources away from core activities.
“By reallocating these funds toward the development and commercialization of AI technologies, BenevolentAI can more effectively meet partners’ needs,” the company said in its announcement. “Additionally, transitioning to a private company would grant greater operational agility and reduced corporate complexity, enabling the Company to better lead the dynamic TechBio sector it has helped to pioneer and develop over the last decade.”
Illustration: metamorworks, Getty Images
高管变更财报临床2期
2024-12-11
Today, a brief rundown of news involving BenevolentAI and Tasca Therapeutics, as well as updates from Eli Lilly, AbbVie, Biogen that you may have missed.BenevolentAI co-founder Kenneth Mulvaney says he is taking the AI drug discovery specialist back to its techbio foundation in a restructuring announced by the firm Wednesday. Mulvaney returned to lead BenevolentAI in October, replacing then-CEO Joerg Moeller. Reshaping BenevolentAI involved tough but necessary decisions, Mulvaney said in a statement that outlined BenevolentAIs next steps, which include layoffs and a plan to delist from the Euronext Amsterdam stock exchange. The company also said that, going forward, it would attempt to partner its drug candidates earlier in development to reduce its balance sheet risk. The Board is wholly behind this strategic direction and restoration of the business to its TechBio roots, deputy chair Peter Allen said in the statement. Ned PagliaruloTasca Therapeutics raised $52 million in a Series A round, the biotechnology startup said Tuesday. Backed by Cure Ventures and Regeneron Ventures, the company claims it has a new way to identify cancer drug candidates. The Series A will be used to fund development of its lead program, called CP-383, in Phase 1/2 studies in people with cancers of the lung, colon, head and neck, and brain. Gwendolyn WuEli Lillys board of directors has given the company a green light to buy back up to $15 billion worth of stock, triple the size of Lillys last share repurchase program. The board also authorized a 15% increase to Lillys dividend. Given the strong growth profile of the company, we're ... increasing the amount of capital we plan to return to shareholders, Lilly CFO Lucas Montarce said in a Monday statement. Lilly shares are up about 37% this year, although its stock price growth has cooled in recent months. The company plans to complete its newly authorized share buybacks within three years. Ned PagliaruloAn experimental drug for Parkinsons disease has succeeded in a second late-stage clinical trial, convincing its owner, AbbVie, theres enough evidence to file for approval. While few details were given, AbbVie said the drug, called tavapadon, was significantly better than a placebo at improving patients ability to do daily tasks that require muscle movement, like speaking, eating and writing. The company plans to submit an approval application sometime next year. Tavapadon was one of the assets AbbVie got last year through its nearly $9 billion acquisition of Cerevel Therapeutics. Another key asset from that deal, the schizophrenia drug emraclidine, suffered a major setback last month. Jacob BellBiogen has poached a big pharma investor whisperer, announcing Monday the appointment of Tim Power as head of investor relations. Its the latest addition to an executive team that has undergone considerable change over the past couple years. Power spent the better part of the last decade dealing with investors for Bristol Myers Squibb. Now, at Biogen, he will report to Chief Financial Officer Michael McDonnell, who is set to retire in February. McDonnell said in a statement that Powers expertise, communication skills and strong background in articulating breakthrough scientific advancements will be invaluable for Biogen. Jacob Bell '
高管变更并购IPO
100 项与 BenevolentAI Ltd. 相关的药物交易
登录后查看更多信息
100 项与 BenevolentAI Ltd. 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年04月04日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
药物发现
2
2
临床前
临床1期
1
1
临床2期
其他
13
登录后查看更多信息
当前项目
| 药物(靶点) | 适应症 | 全球最高研发状态 |
|---|---|---|
BEN-2293 ( NTRK x TrkA x TrkB x TrkC ) | 中度特应性皮炎 更多 | 临床1/2期 |
BEN-8744 ( PDE10A ) | 溃疡性结肠炎 更多 | 临床前 |
BEN-34712 ( RARα x RARβ2 ) | 肌萎缩侧索硬化 更多 | 临床前 |
BEN-28010 ( Chk1 ) | 胶质母细胞瘤 更多 | 临床前 |
Fibrosis therapy(BenevolentAI Ltd) | 纤维化 更多 | 药物发现 |
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
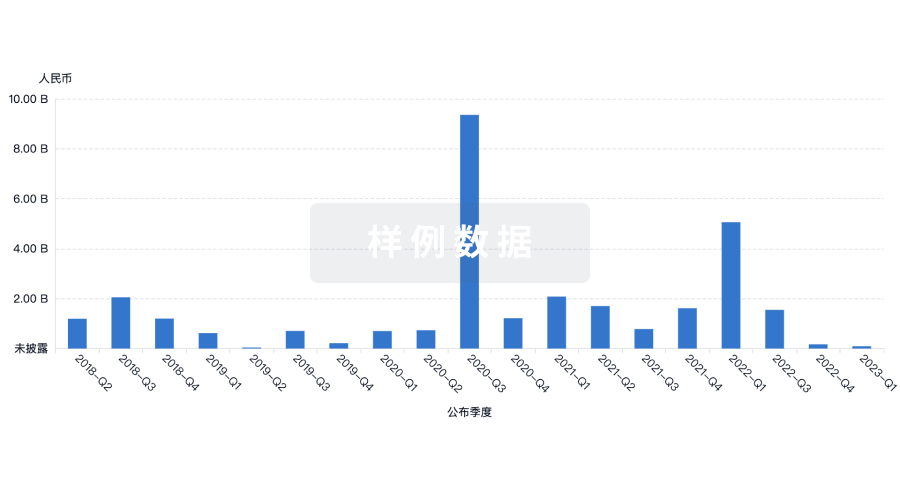
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
来和芽仔聊天吧
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用