预约演示
更新于:2025-08-29

The University of Suwon
更新于:2025-08-29
概览
关联
100 项与 The University of Suwon 相关的临床结果
登录后查看更多信息
0 项与 The University of Suwon 相关的专利(医药)
登录后查看更多信息
1,011
项与 The University of Suwon 相关的文献(医药)2025-09-01·WATER RESEARCH
Decay and solid-liquid partitioning of mpox and vaccinia virus DNA in primary influent and settled solids to guide wastewater-based epidemiology practices
Article
作者: Graham, Katherine E ; Cha, Gyuhyon ; Phaneuf, Jacob R ; Konstantinidis, Konstantinos T ; Hatt, Janet K
Wastewater-based epidemiology (WBE) has proven to be a powerful tool for tracking the spread of viral pathogens, such as SARS-CoV-2; however, as WBE has expanded to include new pathogens, like mpox virus (MPXV), more data is needed to guide practitioners on how to design WBE campaigns. Here, we investigated the decay rates of heat-inactivated MPXV (HI-MPXV) and attenuated vaccinia virus (VV) viral signal in primary influent and settled solids collected from a local POTW at 4 °C, 22 °C, or 35 °C using digital PCR. Subsequently, we studied the solid-liquid partitioning of the viruses in primary influent. Over 30 days, decay rates did not significantly differ between viruses (p = 0.5258). However, decay was significantly higher in primary influent (0.109-0.144/day) than in settled solids (0.019-0.040/day) at both 22 °C (p = 0.0030) and 35 °C (p = 0.0166). Furthermore, as part of the partitioning experiment, we found that HI-MPXV and VV adsorb to the solids fraction of primary influent at higher intensities than previously studied enveloped viruses (KF = 1000-31,800 mL/g, n = 1.01-1.41). Likewise, it was determined in the partitioning experiment that a concentration of greater than 103 gc/mL in raw influent was needed for the viable quantification of MPXV and VV DNA in the clarified liquid fraction of raw primarily influent. Our study provides essential insights into informative sample collection and storage conditions for the analysis of wastewater and for transport modeling studies. Due to the slow decay observed in settled solids at all tested temperatures in the persistence experiment, this matrix may be most suitable for retrospective analyses of MPXV infections within a community.
2025-09-01·WATER RESEARCH
Genomic insights into chlorine resistance of a Mycobacterium sp. strain isolated from treated wastewater effluent
Article
作者: Matsuura, Norihisa ; Konstantinidis, Konstantinos T ; Galagoda, Rasindu ; Cha, Gyuhyon ; Honda, Ryo ; Hara-Yamamura, Hiroe
Chlorine is the principal microbial disinfectant used for water treatment. However, chlorine-resistant bacteria such as Mycobacterium spp., can survive chlorine treatment and even grow in the presence of chlorine, posing potential public health risks. In this study, we isolated a Mycobacterium sp. strain from treated effluent and investigated its chlorine resistance and recovery using transcriptomic analyses. Specifically, isolate M1, showing 94.58 % average nucleotide identity (ANI) with Mycobacterium massilipolynesiensis type genome, was exposed to 1 ppm HOCl for 30 min and subjected to RNA sequencing. Genes identified as upregulated compared to control conditions (no HOCl) were involved in detoxification (toxic compound degradation; nemA; log2 fold-change [FC]: 7.41), redox homeostasis (COQ5; quinone synthesis; log2 FC 5.70, rosB; riboflavin synthesis; log2 FC 5.61), protein homeostasis (cysHKO, moeZ cysteine biosynthesis, and arg complex; arginine metabolism), and lipid metabolism (cpnA; 6.95 FC) suggesting a multifaceted adaptation to oxidative stress. Levels of a few membrane transport proteins (czcD, and bcr) were also upregulated, highlighting their role in chlorine exposure. Overall, this study broadens the understanding of chlorine resistance strategies employed by Mycobacterium sp. to combat oxidative stress and the resulting toxic intracellular compounds, and has implications for adjusting water treatment technologies toward eliminating mycobacteria.
2025-08-20·ACS Applied Materials & Interfaces
Nonthermal Atmospheric Plasma Induces Osteogenic Activity in Osteoblasts through the ROS/WNT Signaling Axis
Article
作者: Choi, Eun Ha ; Sharma, Garima ; Kaushik, Neha ; Acharya, Tirtha Raj ; Jagga, Supriya ; Sharma, Ashish Ranjan ; Kaushik, Nagendra Kumar ; Lee, Sang-Soo ; Kim, Jin-Chul
Nonthermal plasma (NTP) has garnered attention for its potential in various biological activities. Due to the interactions with proteins and other cellular components, NTPs might stimulate osteoblast activity, but the exact mechanisms are still unknown. This study used two kinds of NTP: dielectric barrier discharge (DBD) and jet NTP (JNTP) to treat serum-free cell culture media at different time intervals. The indirect (plasma-media interfaces of ion exchange) exposure to osteoblasts was analyzed to determine whether it could influence osteoblast behavior and stimulate their osteogenic activity. Among various NTP-ionized media, 25% to 100% of the JNTP medium (JNTPM, treated for 10 min) showed noticeable induction of ALP activity in osteoblasts. 25% JNTPM induced maximum ALP activity and also demonstrated enhanced mRNA expression of osteogenic markers like Runx-2, Osterix, collagen 1α, bone sialoprotein, and osteocalcin. In addition, JNTPM treatment to osteoblasts demonstrated enhanced collagen production and mineralization. The administration of JNTPM to SaOS-2 cells increased Axin-2 reporter activity, along with enhanced stabilization of β-catenin and phosphorylation of GSK3-β, indicating the stimulation of the WNT signaling pathway. Moreover, JNTPM treatment increased osteoblasts' intracellular reactive oxygen species (ROS) levels. Prior exposure of N-acetyl-l-cysteine (NAC) to JNTPM-treated osteoblasts diminished the phosphorylation of GSK3-β, Axin-2 reporter activity, and the stability of β-catenin, highlighting the importance of intracellular ROS in enhancing Wnt signaling and osteogenic activity in osteoblasts in response to JNTPM. Thus, JNTPM treatment induces osteogenic stimulatory properties in osteoblasts through cross-talk between ROS and the Wnt signaling axis, suggesting a potential therapeutic approach for bone formation.
100 项与 The University of Suwon 相关的药物交易
登录后查看更多信息
100 项与 The University of Suwon 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年10月20日管线快照
无数据报导
登录后保持更新
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
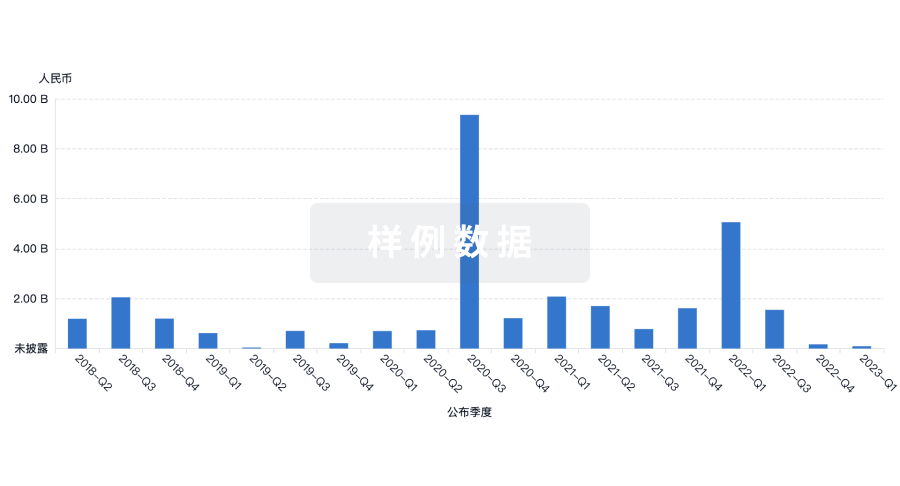
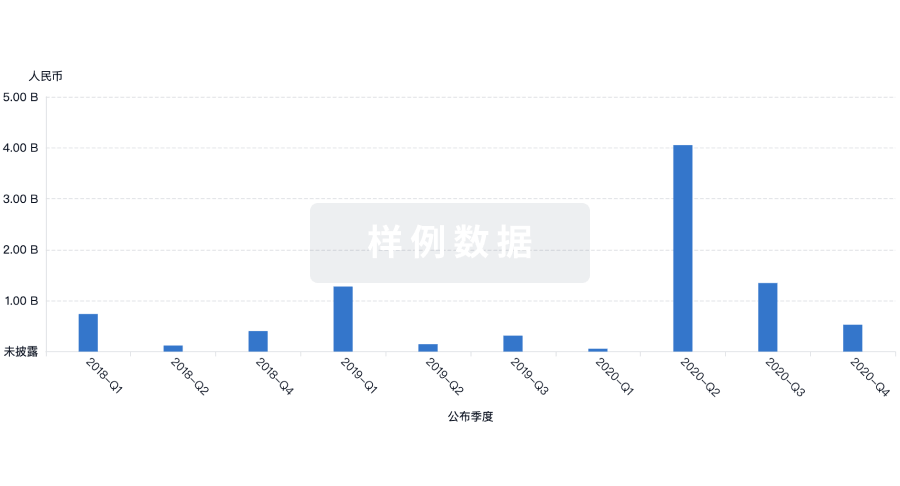
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用