预约演示
更新于:2025-05-07
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
更新于:2025-05-07
概览
关联
280
项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的临床试验NCT06950255
Effect of a 3-week Program of Cane Training and Use on Gait of Individuals With Parkinson's Disease: Protocol for a Randomized Controlled Trial
Introduction: Although individuals with Parkinson's disease (PD) commonly use assistive devices, the effects of these devices on gait remain poorly understood. Furthermore, previous studies on this topic only investigated the immediate effects of device usage, and the investigated outcomes were predominately performance-based neglecting participant-centered outcomes.
Objective: To investigate the effect of cane training and use on gait speed (primary outcome), gait confidence, cadence, step length, functional mobility, freezing of gait, fear of falls and satisfaction with the use of a cane (secondary outcomes) in individuals with PD.
Methods: A double-blind, randomized controlled trial with intention-to-treat and per-protocol analysis will be carried out. A total of 26 individuals with PD will be recruited based on the following inclusion criteria: age ≥ 40 years, diagnosis of idiopathic PD, classification between stages II to IV on the modified Hoehn & Yahr Scale, stable use of anti-parkinsonian pharmacological therapy, ability to walk independently with a walking speed ≤ 1.1 m/s (defined to screen individuals with gait impairments), and ability to use a single-point cane during walking without regular use of any type of assistive device since the diagnosis of PD. Participant will be randomly divided into two groups that will receive: (1) cane training and use (experimental group) or (2) global stretches and health education (time and attention-controlled group). The intervention will be provided in four sessions lasting 40 minutes each, spaced over 15 to 22 days. Additionally, individuals will be instructed to use a cane (experimental group) or perform stretching exercises (time and attention-controlled group) daily, starting from the first day of training. Assessments will be conducted at the beginning of the study (week 0), post-intervention (after the 3-week intervention), and one month after the cessation of the intervention (8-week follow-up). The primary outcomes is gait speed. Secondary outcomes include gait confidence, cadence, step length, functional mobility, freezing of gait, fear of falls, and satisfaction with the use of a cane. Between-group differences will be measured using a two-way repeated measures ANOVA, considering baseline, post-intervention, and follow-up assessments, following both intention-to-treat and per-protocol approaches (α=0.05).
Conclusions: The results of the present study will provide information about the effects of cane training and use on the gait of individuals with PD who will have the opportunity to use the cane in their real-life context.
Objective: To investigate the effect of cane training and use on gait speed (primary outcome), gait confidence, cadence, step length, functional mobility, freezing of gait, fear of falls and satisfaction with the use of a cane (secondary outcomes) in individuals with PD.
Methods: A double-blind, randomized controlled trial with intention-to-treat and per-protocol analysis will be carried out. A total of 26 individuals with PD will be recruited based on the following inclusion criteria: age ≥ 40 years, diagnosis of idiopathic PD, classification between stages II to IV on the modified Hoehn & Yahr Scale, stable use of anti-parkinsonian pharmacological therapy, ability to walk independently with a walking speed ≤ 1.1 m/s (defined to screen individuals with gait impairments), and ability to use a single-point cane during walking without regular use of any type of assistive device since the diagnosis of PD. Participant will be randomly divided into two groups that will receive: (1) cane training and use (experimental group) or (2) global stretches and health education (time and attention-controlled group). The intervention will be provided in four sessions lasting 40 minutes each, spaced over 15 to 22 days. Additionally, individuals will be instructed to use a cane (experimental group) or perform stretching exercises (time and attention-controlled group) daily, starting from the first day of training. Assessments will be conducted at the beginning of the study (week 0), post-intervention (after the 3-week intervention), and one month after the cessation of the intervention (8-week follow-up). The primary outcomes is gait speed. Secondary outcomes include gait confidence, cadence, step length, functional mobility, freezing of gait, fear of falls, and satisfaction with the use of a cane. Between-group differences will be measured using a two-way repeated measures ANOVA, considering baseline, post-intervention, and follow-up assessments, following both intention-to-treat and per-protocol approaches (α=0.05).
Conclusions: The results of the present study will provide information about the effects of cane training and use on the gait of individuals with PD who will have the opportunity to use the cane in their real-life context.
开始日期2025-05-01 |
申办/合作机构 |
NCT06646523
Effects of Remotely Supervised Home-based High-speed Bodyweight Resistance Training on Bradykinesia in Individuals with Parkinson's Disease: a Randomized Clinical Trial
Exercises that involve increasing the speed of movements are beneficial for individuals with Parkinson's disease (PD) and have the potential to reduce bradykinesia and improve mobility. High-speed bodyweight resistance training is a treatment that involves increasing speed considered accessible and viable as it can be performed at any time and place, including at home. This treatment has already shown benefits in the elderly individuals, however no studies were found in individuals with PD. Therefore, the primary aim of this study will be to investigate the effects of home-based and remotely supervised high-speed bodyweight resistance training in reducing bradykinesia in individuals with PD. The secondary aim will be to investigate the effects of home-based and remotely supervised high-speed bodyweight resistance training in improving mobility, muscle power, dynamic balance, and quality of life in this population. A randomized controlled trial will be carried out with concealed allocation, blinded assessments, and intention-to-treat analysis. Altogether, 46 individuals with PD (age ≥ 50 years old, who are bradykinetics and sedentary or insufficiently active will be included. Participants will be randomly assigned to either an experimental group (high-speed bodyweight resistance training) or a control group (bodyweight intervention, usual speed). Both groups will perform a home-based and remotely supervised intervention, consisting of 60-min individual sessions, three times per week over 12 weeks, with a trained physiotherapist. Primary outcomes are bradykinesia of the lower limbs and mobility. Secondary outcomes are muscle power, dynamic balance, and quality of life. Between-group differences will be measured by two-way repeated measures ANOVA, considering the baseline, post-training, and 4-week follow-up. The findings of this trial have the potential to provide important insights regarding the effects of high-speed bodyweight resistance training in reducing bradykinesia and improving mobility in individuals with PD. High-speed bodyweight resistance training does not use any type of external resistance and can be performed anywhere and at any time. In addition, it can be performed at home through telemonitoring, reducing time and costs of transport, making it quite feasible and accessible for individuals from different social and economic backgrounds which increases the feasibility of reproducing their findings in clinical practice.
开始日期2024-10-18 |
申办/合作机构 |
NCT06288204
Association of Green Propolis Extract and Royal Jelly to Modulate Inflammation and Oxidative Stress in Hypertensive Patients and/or With Chronic Kidney Disease
This work aims to evaluate the effects of the association of green propolis extract with royal jelly on inflammation and oxidative stress in participants with chronic kidney diseases (CKD) and Systemic arterial hypertension (SAH), in a longitudinal, randomized, double-blind, placebo-controlled clinical trial that will be carried out for 2 months.
开始日期2024-10-01 |
申办/合作机构 |
100 项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的临床结果
登录后查看更多信息
0 项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的专利(医药)
登录后查看更多信息
39
项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的文献(医药)2022-07-20·International Journal of Systematic and Evolutionary Microbiology
Bradyrhizobium cenepequi sp. nov., Bradyrhizobium semiaridum sp. nov., Bradyrhizobium hereditatis sp. nov. and Bradyrhizobium australafricanum sp. nov., symbionts of different leguminous plants of Western Australia and South Africa and definition of three novel symbiovars
Article
作者: Hungria, Mariangela ; Helene, Luisa Caroline Ferraz ; O´Hara, Graham ; Klepa, Milena Serenato
2022-05-21·International Journal of Microbiology
New Insights into the Taxonomy of Bacteria in the Genomic Era and a Case Study with Rhizobia
Review
作者: Klepa, Milena Serenato ; Hungria, Mariangela ; Ferraz Helene, Luisa Caroline
2021-10-01·Archives of Microbiology4区 · 生物学
Twenty years of paradigm-breaking studies of taxonomy and symbiotic nitrogen fixation by beta-rhizobia, and indication of Brazil as a hotspot of Paraburkholderia diversity
4区 · 生物学
Review
作者: Hungria, Mariangela ; Paulitsch, Fabiane ; Dos Reis, Fabio Bueno
100 项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的药物交易
登录后查看更多信息
100 项与 Coordenação de Aperfeiçoamento de Pessoal de Nível Superior 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2025年09月22日管线快照
无数据报导
登录后保持更新
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
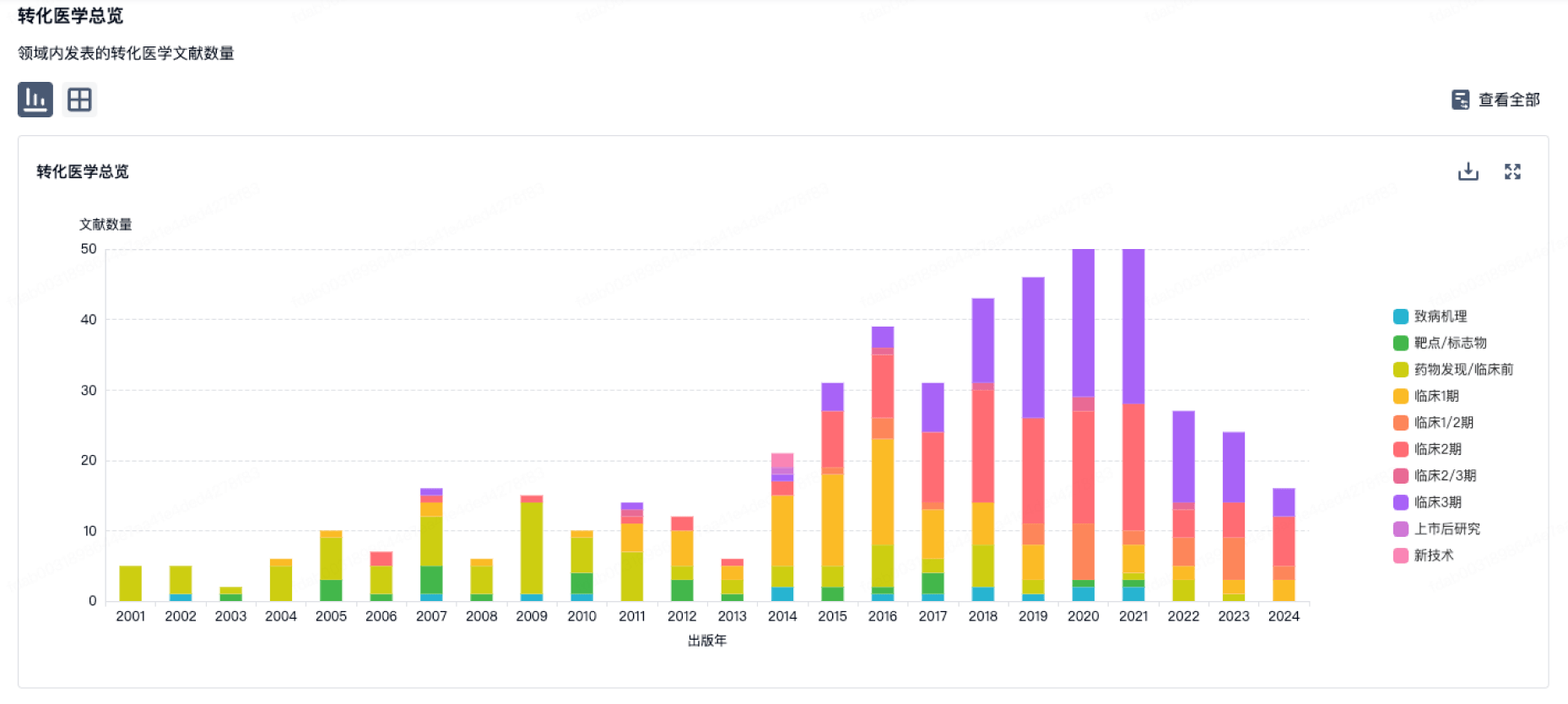
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
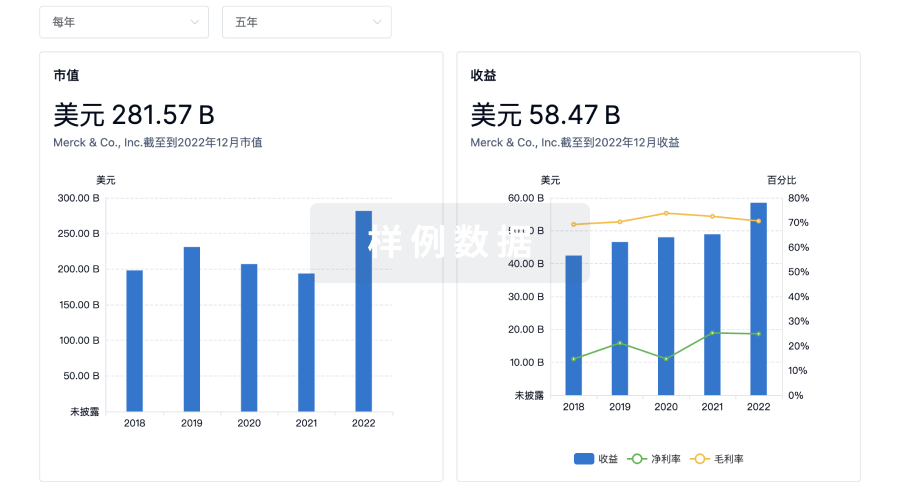
营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或
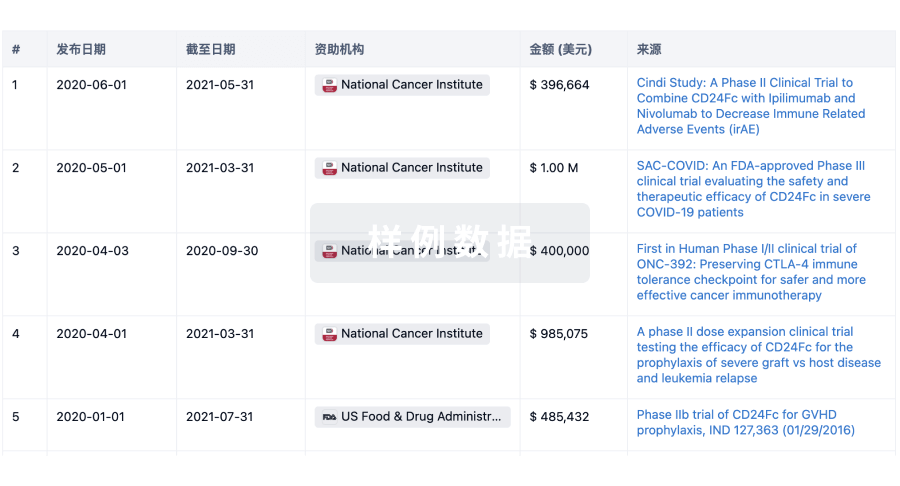
科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或
投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或
融资
发掘融资趋势以验证和推进您的投资机会。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

