预约演示
更新于:2025-08-02
MT104
更新于:2025-08-02
概要
基本信息
原研机构 |
在研机构 |
非在研机构- |
权益机构- |
最高研发阶段临床前 |
首次获批日期- |
最高研发阶段(中国)- |
特殊审评- |
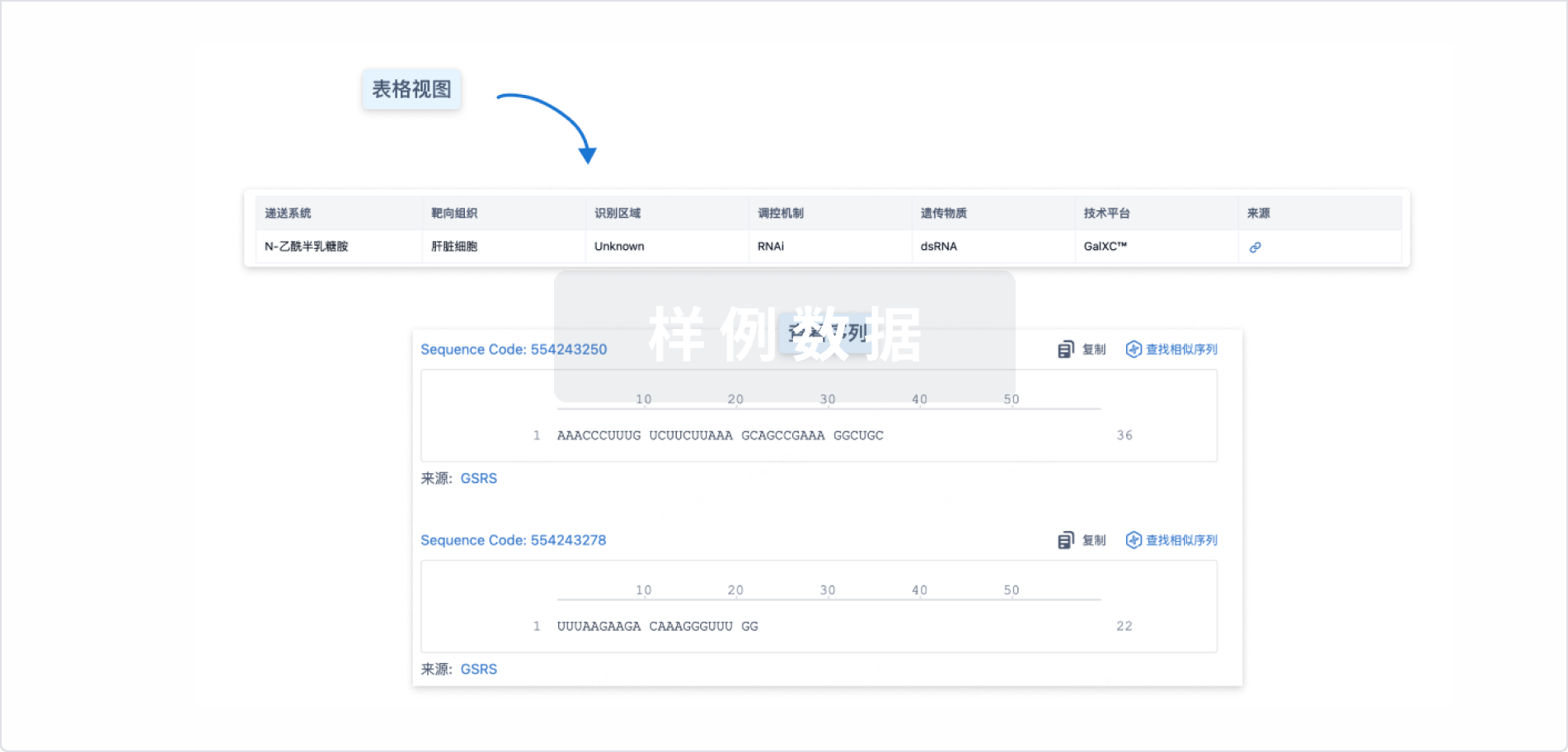
结构/序列
使用我们的RNA技术数据为新药研发加速。
登录
或
关联
1
项与 MT104 相关的临床试验KCT0006499
Single center, randomized, double-blind, placebo-controlled clinical trial for evaluation of efficacy on cognitive function and safety of MT104
开始日期2021-07-01 |
100 项与 MT104 相关的临床结果
登录后查看更多信息
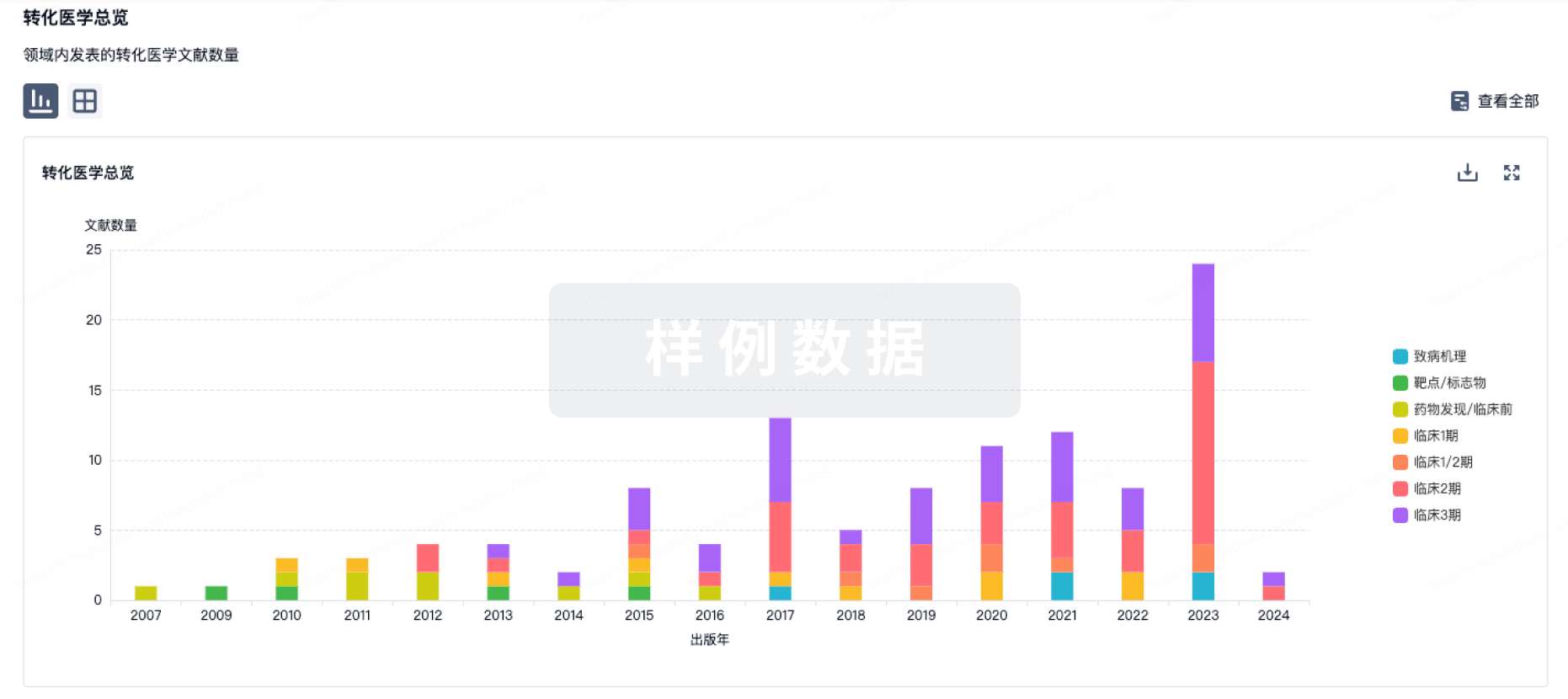
100 项与 MT104 相关的转化医学
登录后查看更多信息
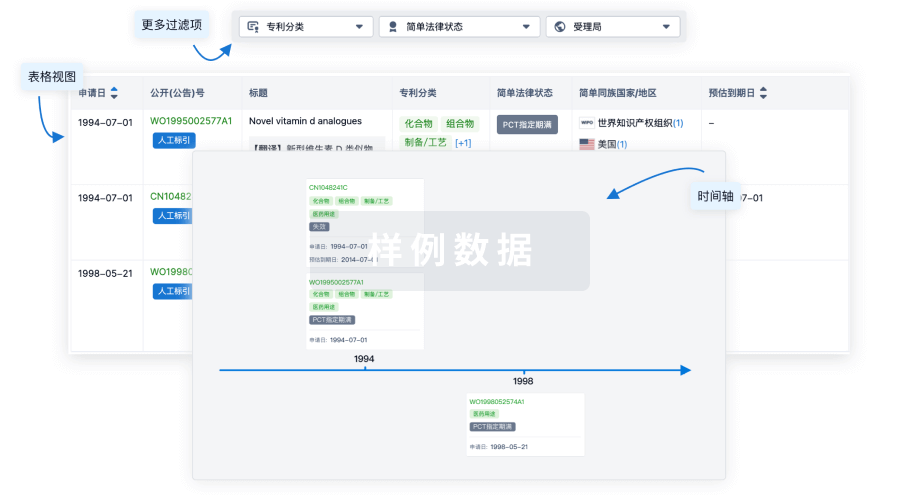
100 项与 MT104 相关的专利(医药)
登录后查看更多信息
3
项与 MT104 相关的文献(医药)2005-06-01·Carbohydrate research4区 · 化学
The structure of the antigenic O-polysaccharide of the lipopolysaccharide of Edwardsiella ictaluri strain MT104
4区 · 化学
Article
作者: William W. Kay ; Malcolm B. Perry ; Ludmila Nossova ; Evgeny Vinogradov
The structural characterization of the antigenic O-polysaccharide component of the lipopolysaccharide produced by the fish pathogenic bacterium Edwardsiella ictaluri MT104 was undertaken by the application of NMR spectroscopy and chemical analysis. The O-chain was found to be a linear polymer of a repeating tetrasaccharide unit composed of D-glucose, 2-acetamido-2-deoxy-D-galactose, and D-galactose in a 1:2:1 ratio having the structure: [carbohydrate structure]; see text.
2005-03-01·International journal of systematic and evolutionary microbiology3区 · 生物学
Novosphingobium lentum sp. nov., a psychrotolerant bacterium from a polychlorophenol bioremediation process
3区 · 生物学
Article
作者: Busse, Hans-Jürgen ; Tiirola, Marja A. ; Kämpfer, Peter ; Männistö, Minna K.
A polychlorophenol-degrading strain, designated MT1T, and three MT1-like strains, MT101, MT103 and MT104, were isolated from a cold (4–8 °C) fluidized-bed process treating chlorophenol-contaminated groundwater in southern Finland. The organisms were Gram-negative, rod-shaped, catalase-positive, non-spore-forming and non-motile. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the strains belonged to theα-4 subclass of theProteobacteriaand were members of the genusNovosphingobium. The highest 16S rRNA gene sequence similarity observed for these strains was 96·5 % with the type strains ofNovosphingobium hassiacum,Novosphingobium aromaticivoransandNovosphingobium subterraneum. Chemotaxonomic data (major ubiquinone: Q-10; major polyamine: spermidine; major polar lipids: phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylcholine and sphingoglycolipid; major fatty acids: 18 : 1ω7c, 16 : 1ω7cand 2-OH 14 : 0) as well as the ability to reduce nitrate supported the affiliation of the strains to the genusNovosphingobium. Based on the phylogenetic analysis, whole-cell fatty acid composition as well as biochemical and physiological characteristics, the MT1-like strains were highly similar and could be separated from all recognizedNovosphingobiumspecies. The novel speciesNovosphingobium lentumsp. nov. is proposed to accommodate strains MT1T(=DSM 13663T=CCUG 45847T), MT101 (=CCUG 45849), MT103 (=CCUG 45850) and MT104 (=CCUG 45851).
JOURNAL OF MEDICINAL FOOD
Efficacy and Safety of MT104, A Dietary Supplement Based on Cuscuta Seeds and Heat-Killed Probiotics, on Cognitive Function in Patients with Mild Cognitive Impairment: A 12-Week, Multicenter, Randomized, Double-Blind, Placebo-Controlled Clinical Trial
Article
作者: Choi, Sang Zin ; Kim, Bo-Hyung ; Lee, Chan-Nyoung ; Kwon, Minji ; Park, Jin-Woo ; Lee, Jin San ; Chae, Soo-Wan ; Park, Sang Cheol ; Sohn, Mi Won ; Yim, Sung-Vin ; Choi, Jin Gyu
Mild cognitive impairment (MCI) represents the symptomatic predementia stage of Alzheimer's disease (AD). To delay the progression of MCI to AD, appropriate interventions capable of modulating the microbiota-gut-brain (MGB) axis are necessary. This study was designed to assess the efficacy and safety of MT104, a dietary supplement comprising Cuscuta seeds and heat-killed probiotics, based on the biological mechanisms regulating the MGB axis in patients with MCI. This was a multicenter, randomized, double-blind, and placebo-controlled study. All patients were randomly allocated in a 1:1 ratio to the MT104 or placebo group. Global cognition was assessed using the Korean-Montreal Cognitive Assessment (K-MoCA) and Korean-Mini Mental State Examination at baseline and after 12 weeks of treatment. The visuospatial function was assessed using the copying performance from the Rey Complex Figure Test (RCFT), and verbal and visual memory functions were evaluated using the Seoul Verbal Learning Test (SVLT) and RCFT. Differences between groups were analyzed using either the t-test or the Mann-Whitney U test. Analyses of covariance and ranked analysis of covariance were performed. The mean changes in verbal memory function, as measured by the SVLT delayed recall, showed clinically significant improvement in the MT104 group relative to the placebo group in the intention-to-treat and per-protocol groups. Global cognition, as measured using the K-MoCA, also significantly improved in the per-protocol group. In addition, no significant findings were identified. This study highlighted the potential of dietary therapeutic strategies focused on reducing the risk of progression from MCI to AD.
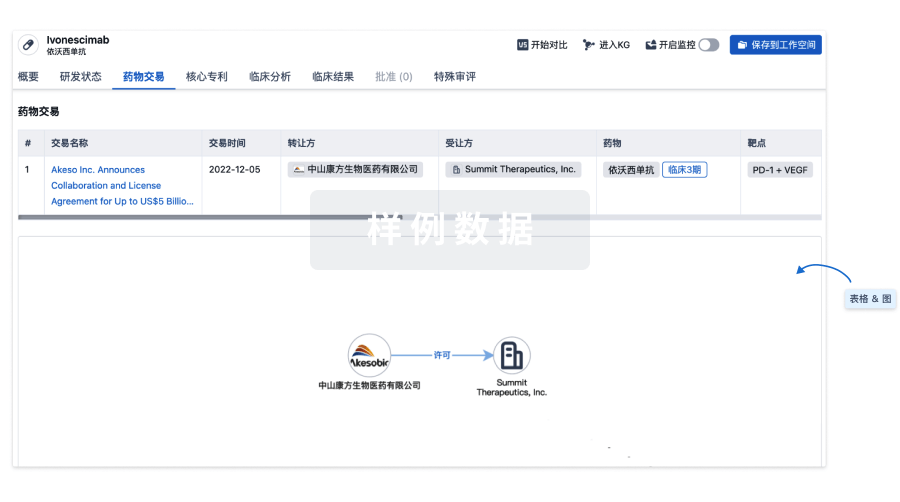
100 项与 MT104 相关的药物交易
登录后查看更多信息
研发状态
10 条进展最快的记录, 后查看更多信息
登录
| 适应症 | 最高研发状态 | 国家/地区 | 公司 | 日期 |
|---|---|---|---|---|
| 阿尔茨海默症 | 临床前 | 韩国 | 2023-04-25 | |
| 认知功能障碍 | 临床前 | 韩国 | 2023-04-25 |
登录后查看更多信息
临床结果
临床结果
适应症
分期
评价
查看全部结果
| 研究 | 分期 | 人群特征 | 评价人数 | 分组 | 结果 | 评价 | 发布日期 |
|---|
No Data | |||||||
登录后查看更多信息
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
核心专利
使用我们的核心专利数据促进您的研究。
登录
或
临床分析
紧跟全球注册中心的最新临床试验。
登录
或
批准
利用最新的监管批准信息加速您的研究。
登录
或
特殊审评
只需点击几下即可了解关键药物信息。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

