预约演示
更新于:2025-06-28
TIGIT(Abzyme Therapeutics)
更新于:2025-06-28
概要
基本信息
非在研机构- |
权益机构- |
最高研发阶段临床前 |
首次获批日期- |
最高研发阶段(中国)- |
特殊审评- |
关联
100 项与 TIGIT(Abzyme Therapeutics) 相关的临床结果
登录后查看更多信息
100 项与 TIGIT(Abzyme Therapeutics) 相关的转化医学
登录后查看更多信息
100 项与 TIGIT(Abzyme Therapeutics) 相关的专利(医药)
登录后查看更多信息
46
项与 TIGIT(Abzyme Therapeutics) 相关的文献(医药)2025-10-01·COMPUTATIONAL BIOLOGY AND CHEMISTRY
Pinpointing potent hits for cancer immunotherapy targeting the TIGIT/PVR pathway using the XGBoost model, centroid-based virtual screening, and MD simulation
Article
作者: Jin, Yuanyuan ; Fan, Shuai ; Lü, Xudong ; Li, Jing ; Xia, Zhiyong ; Tang, Mengjia ; Yang, Zhaoyong ; Wang, Chenyu
T cell immunoreceptor with immunoglobulin and ITIM domain (TIGIT) is one of the most promising targets for cancer immunotherapy. The combination of TIGIT and poliovirus receptor (PVR), which is highly expressed on the tumor surface, inhibits the killing of tumor cells by immune cells. Although antibody blocking the PVR/TIGIT immune checkpoint has shown encouraging anti-tumor effects, small molecules targeting TIGIT to block PVR/TIGIT have not yet been studied. In this study, diverse computational approaches were employed to identify potential inhibitors of this therapeutic targets. First, virtual alanine scanning was used to identify hotspot residues of TIGIT that were effective inhibitory sites. Second, the Extreme Gradient Boosting (XGBoost) classification model and the RO4 rule were used to initially exclude negative compounds. Then, centroid-based virtual screening was used in combination with absorption, distribution, metabolism, excretion, and toxicity (ADMET) prediction to identify the four most promising candidate molecules. Molecular dynamics simulation trajectory analysis showed stable dynamic behavior of the candidate molecules and proteins. Molecular mechanics and Poisson-Boltzmann surface area (MMPBSA) calculations showed that MCULE-5939418698 had the lowest binding free energy (-39.79 kcal/mol). Binding-conformation and energy-decomposition analyses indicated significant involvement of residues L47, Q53, V54 and N58 in inhibitor binding. Principal component analysis and free energy landscape analysis further demonstrated that the binding of MCULE-4861917955 made the system thermodynamically more favorable. Thus, we screened potential inhibitors targeting TIGIT and provide a fresh pipeline for future drug screening research.
2025-04-01·Mucosal Immunology
Gut-homing and intestinal TIGITnegCD38+ memory T cells acquire an IL-12-induced, ex-Th17 pathogenic phenotype in a subgroup of Crohn’s disease patients with a severe disease course
Article
作者: Tuk, Bastiaan ; Aardoom, Martine A. ; Ruemmele, Frank M. ; Joosse, Maria E ; Barendregt, Danielle M.H. ; de Ridder, Lissy ; Tindemans, Irma ; Joosse, Maria E. ; Costes, Lea M.M. ; Croft, Nicholas M. ; Escher, Johanna C ; Kemos, Polychronis ; Costes, Léa M M ; Samsom, Janneke N. ; van Berkel, Lisette A ; Croft, Nicholas M ; Hulleman-van Haaften, Danielle H. ; van Berkel, Lisette A. ; Escher, Johanna C. ; Ruemmele, Frank M ; Calado, Beatriz ; Klomberg, Renz C W ; Hulleman-van Haaften, Daniëlle H ; Barendregt, Daniëlle M H ; Heredia, Maud ; Samsom, Janneke N ; Klomberg, Renz C.W. ; Aardoom, Martine A
CD4+ memory T cell (TM) reactivation drives chronicity in inflammatory bowel disease (IBD), including Crohn's disease (CD) and ulcerative colitis. Defects driving loss of TM regulation likely differ between patients but remain undefined. In health, approximately 40 % of circulating gut-homing CD38+TM express co-inhibitory receptor T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT). TIGIT+CD38+TM have regulatory function while TIGITnegCD38+TM are enriched in IFN-γ-producing cells. We hypothesized TIGITnegCD38+TM are inflammatory and drive disease in a subgroup of IBD patients. We characterized TIGIT+CD38+TM in a uniquely large cohort of pediatric IBD patients from time of diagnosis into adulthood. Circulating TIGITnegCD38+TM frequencies were higher in a subgroup of therapy-naïve CD patients with high plasma IFN-γ and a more severe disease course. TIGITnegCD38+TM were highly enriched in HLA-DR+ and ex-Th17/Th1-like cells, high producers of IFN-γ. Cultures of healthy-adult-stimulated TM identified IL-12 as the only IBD-related inflammatory cytokine to drive the pathogenic ex-Th17-TIGITnegCD38+ phenotype. Moreover, IL12RB2 mRNA expression was higher in TIGITnegCD38+TM than TIGIT+CD38+TM, elevated in CD biopsies compared to controls, and correlated with severity of intestinal inflammation. Overall, we argue that in a subgroup of pediatric CD, increased IL-12 signaling drives reprogramming of Th17 to inflammatory Th1-like TIGITnegCD38+TM and causes more severe disease.
2025-03-12·mBio
Inhibition of TIGIT on NK cells improves their cytotoxicity and HIV reservoir eradication potential
Article
作者: Yu, Xiaowen ; Han, Xiaoxu ; Hu, Qinghai ; Shang, Hong ; Chen, Jiaqi ; Wang, Yue ; Guo, Chenxi ; Zhang, Zining ; Jiang, Yongjun ; Ding, Haibo ; Li, Yidi ; Fu, Yajing
ABSTRACT:
The latent human immunodeficiency virus (HIV) reservoir presents the biggest obstacle to curing HIV chronic infection. Consequently, finding novel strategies to control the HIV reservoir is critical. Natural killer (NK) cells are essential for antiviral immunity. However, the influence of NK cell subsets and their associated inhibitory or activating receptors on their cytotoxicity toward the HIV reservoir has not been fully studied. We investigated the relationship between the percentage of NK cells or NK cell subsets and the HIV reservoir. Our results indicated that the percentage of CD56
−
CD16
+
NK cells was positively associated with HIV reservoir size (i.e., HIV DNA, HIV msRNA, or HIV usRNA). Additionally, we observed that the percentage of IFN-γ
+
NK cells was inversely related to the HIV reservoir. Furthermore, the expression of TIGIT on NK cells, particularly CD56
-
CD16
+
and CD56
dim
NK cell subsets, positively correlated with the HIV reservoir. Notably, individuals with higher percentage of TIGIT
+
NK and lower percentage of CD226
+
NK cells exhibited larger HIV reservoir. Mechanistically, we discovered that TIGIT could inhibit the PI3K-Akt-mTOR-mTORC1 (s6k) signaling pathway to decrease the production of IFN-γ in NK cells. Importantly, inhibiting TIGIT in NK cells enhanced their ability to eliminate reactivated latently infected CD4
+
T cells. Our experiments underscored the crucial role of NK cells in controlling the HIV reservoir and suggested that TIGIT serves as a promising target for enhancing the NK cell-mediated clearance of the HIV reservoir.
IMPORTANCE:
As a major barrier to human immunodeficiency virus (HIV) cure, HIV reservoir persist in viremia-suppressed infected individuals. NK cells are important antiviral cells, and their impact on reservoir has rarely been reported. We analyzed the relationship between the size of reservoir and NK cell subsets, inhibitory receptor TIGIT expression. Our analysis found that the percentage of CD56
−
CD16
+
NK cells was positively associated with HIV reservoir size. Furthermore, TIGIT expression on NK cells and CD56
−
CD16
+
NK cells or CD56
dim
NK cells has a positive correlation with the HIV reservoir. TIGIT can inhibit the PI3K-Akt-mTOR-mTORC1 (s6k) signaling pathway to decrease the production of IFN-γ on NK cells. Blocking TIGIT in NK cells can enhance their ability to eliminate reactivated latently infected CD4
+
T cells. Our study indicated that NK cells are critical to the control of the reservoir size, and TIGIT may be a target for enhancing the NK cell-mediated elimination of the reservoir.
100 项与 TIGIT(Abzyme Therapeutics) 相关的药物交易
登录后查看更多信息
研发状态
10 条进展最快的记录, 后查看更多信息
登录
| 适应症 | 最高研发状态 | 国家/地区 | 公司 | 日期 |
|---|---|---|---|---|
| 肿瘤 | 临床前 | 美国 | 2024-07-16 |
登录后查看更多信息
临床结果
临床结果
适应症
分期
评价
查看全部结果
| 研究 | 分期 | 人群特征 | 评价人数 | 分组 | 结果 | 评价 | 发布日期 |
|---|
No Data | |||||||
登录后查看更多信息
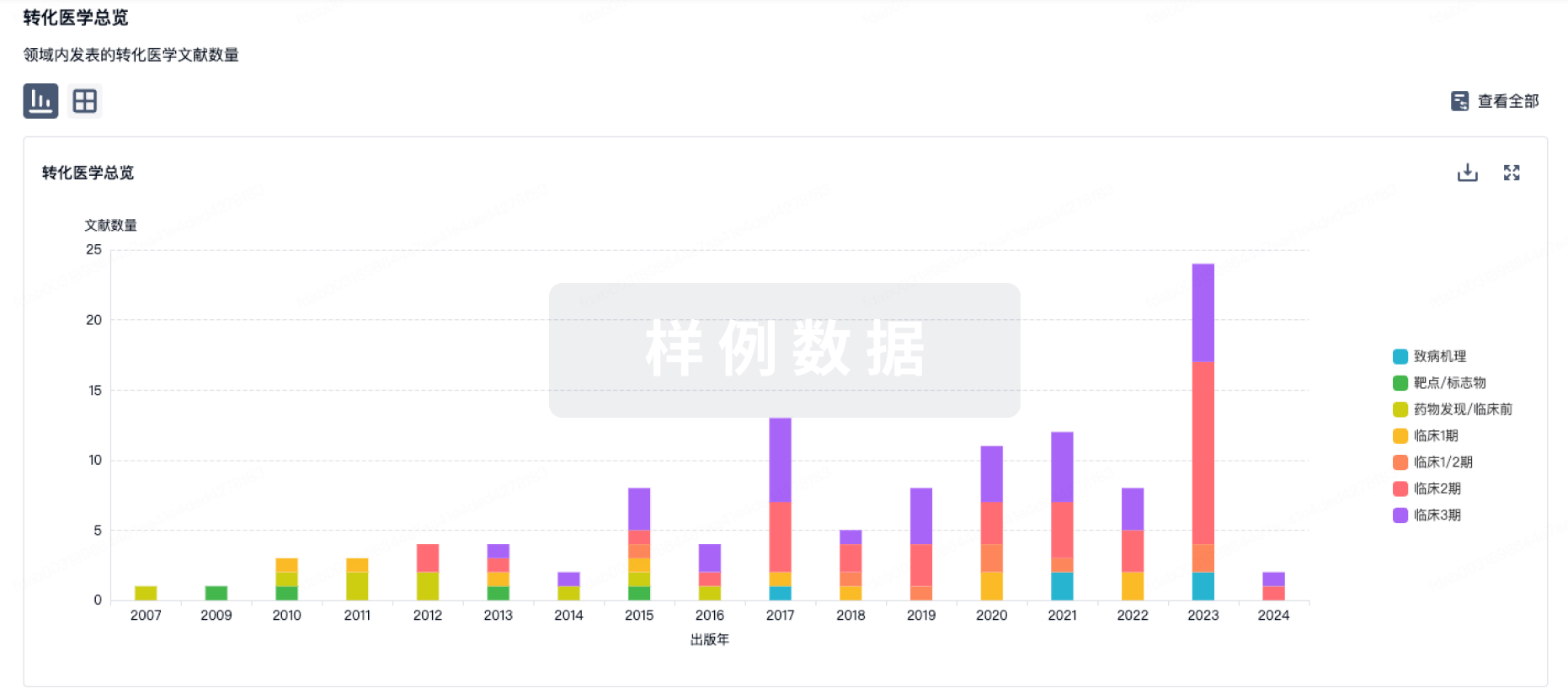
转化医学
使用我们的转化医学数据加速您的研究。
登录
或
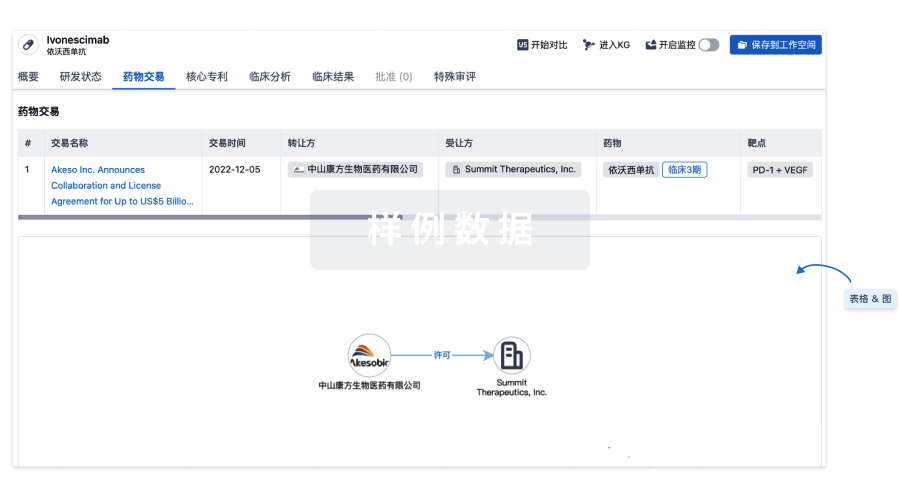
药物交易
使用我们的药物交易数据加速您的研究。
登录
或
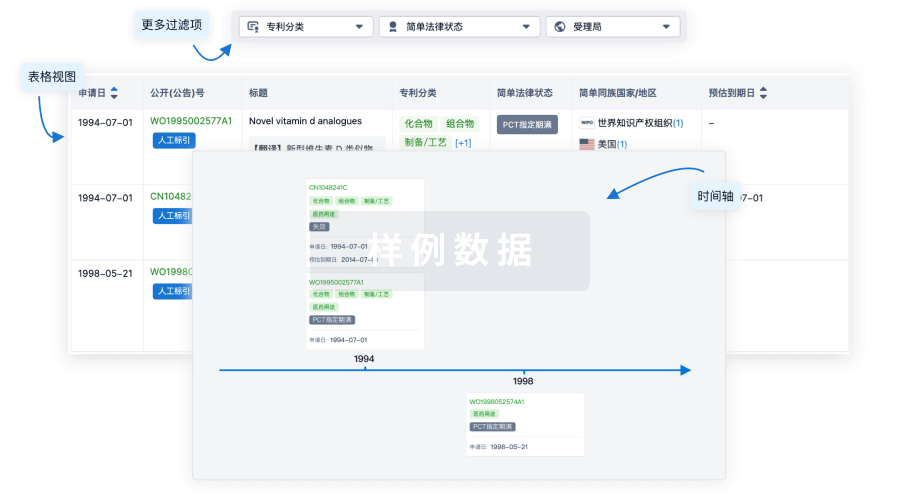
核心专利
使用我们的核心专利数据促进您的研究。
登录
或
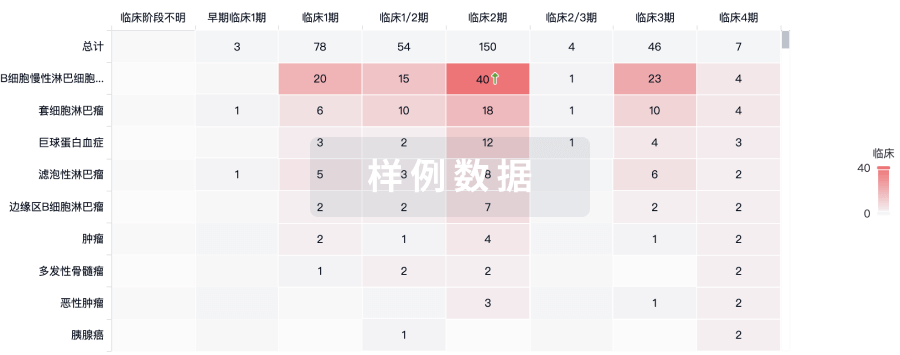
临床分析
紧跟全球注册中心的最新临床试验。
登录
或
批准
利用最新的监管批准信息加速您的研究。
登录
或
生物类似药
生物类似药在不同国家/地区的竞争态势。请注意临床1/2期并入临床2期,临床2/3期并入临床3期
登录
或
特殊审评
只需点击几下即可了解关键药物信息。
登录
或
Eureka LS:
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用
