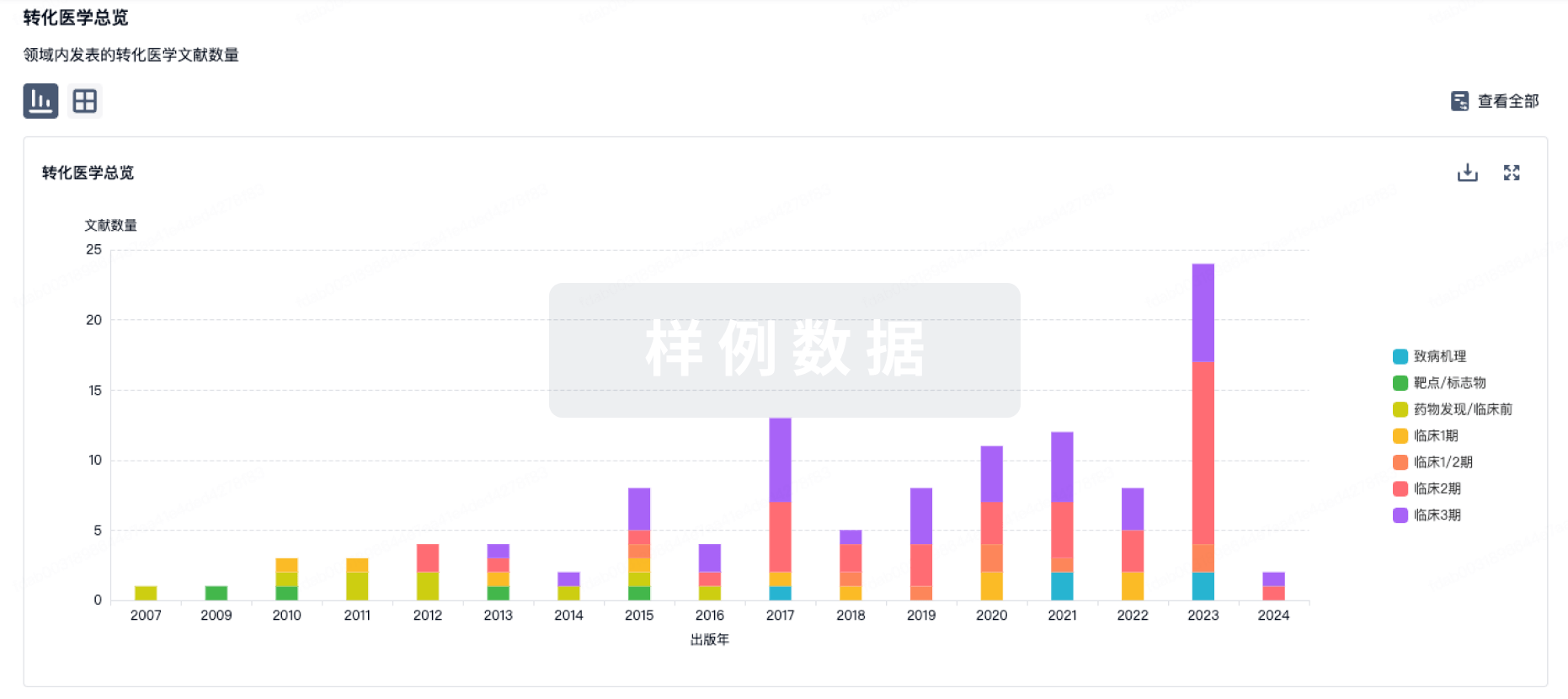
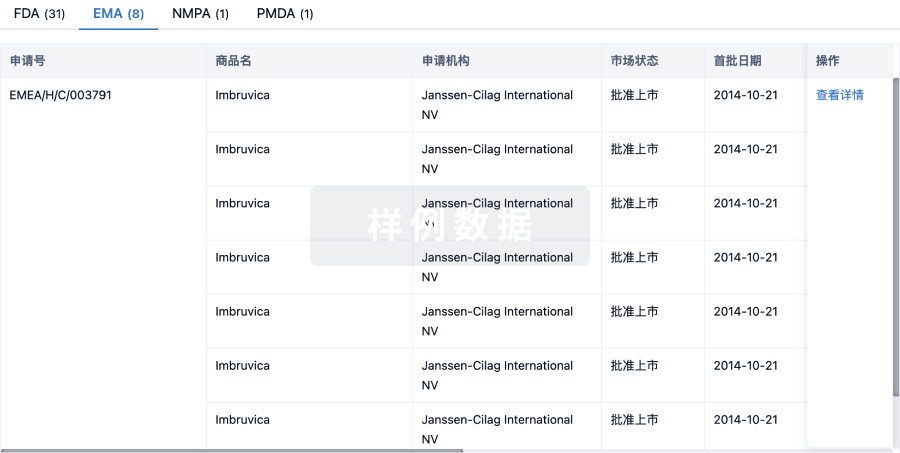
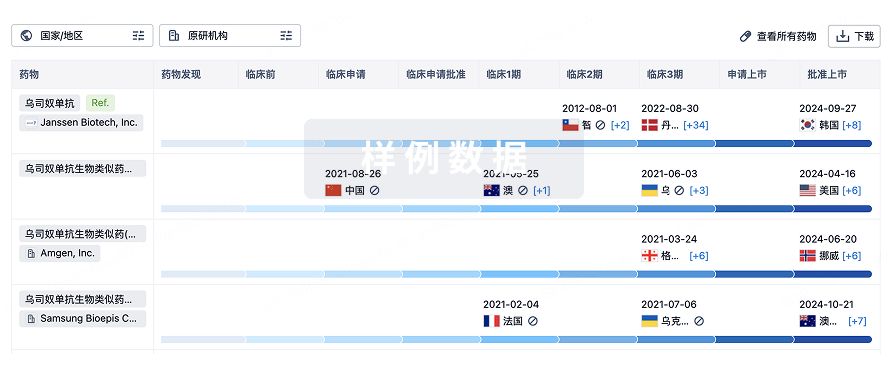
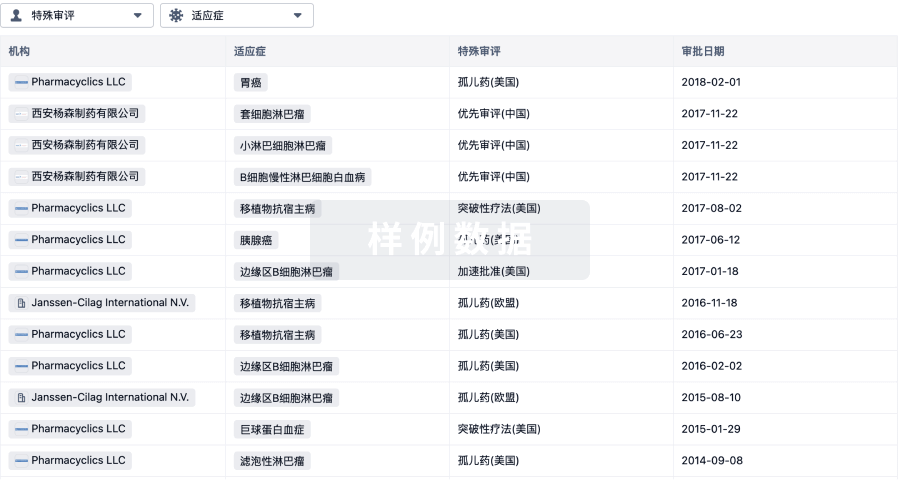
Protein–protein interacting surfaces are usually large and
intricate, making the rational design of small mimetics of these
interfaces a daunting problem. On the basis of a structural similarity
between the CDR2-like loop of CD4 and the β-hairpin region of a short
scorpion toxin, scyllatoxin, we transferred the side chains of nine
residues of CD4, central in the binding to HIV-1 envelope
glycoprotein (gp120), to a structurally homologous region of the
scorpion toxin scaffold. In competition experiments, the resulting
27-amino acid miniprotein inhibited binding of CD4 to gp120 with a 40
μM IC
50
. Structural analysis by NMR showed that both the
backbone of the chimeric β-hairpin and the introduced side chains
adopted conformations similar to those of the parent CD4. Systematic
single mutations suggested that most CD4 residues from the CDR2-like
loop were reproduced in the miniprotein, including the critical Phe-43.
The structural and functional analysis performed suggested five
additional mutations that, once incorporated in the miniprotein,
increased its affinity for gp120 by 100-fold to an IC
50
of
0.1–1.0 μM, depending on viral strains. The resulting mini-CD4
inhibited infection of CD4
+
cells by different virus
isolates. Thus, core regions of large protein–protein interfaces can
be reproduced in miniprotein scaffolds, offering possibilities for the
development of inhibitors of protein–protein interactions that may
represent useful tools in biology and in drug discovery.